A Start Codon Variant in NOG Underlies Symphalangism and Ossicular Chain Malformations Affecting Both the Incus and the Stapes
- PMID: 31428484
- PMCID: PMC6679842
- DOI: 10.1155/2019/2836263
A Start Codon Variant in NOG Underlies Symphalangism and Ossicular Chain Malformations Affecting Both the Incus and the Stapes
Abstract
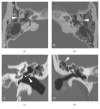
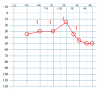
We performed exome sequencing to evaluate the underlying molecular cause of a patient with bilateral conductive hearing loss due to multiple ossicular abnormalities as well as symphalangism of the fifth digits. This leads to the identification of a novel heterozygous start codon variant in the NOG gene (c.2T>C:p.Met1?) that hinders normal translation of the noggin protein. Variants in NOG lead to a spectrum of otologic, digit, and joint abnormalities, a combination suggested to be referred to as NOG-related-symphalangism spectrum disorder (NOG-SSD). Conductive hearing loss from such variants may stem from stapes footplate ankylosis, fixation of the malleoincudal joint, or fixation of the incus short process. In this case, the constellation of both stapes and incus fixation, an exceptionally tall stapes suprastructure, thickened long process of the incus, and enlarged incus body was encountered, leading to distinct challenges during otologic surgery to improve hearing thresholds. This case highlights multiple abnormalities to the ossicular chain in a patient with a start codon variant in NOG. We provide detailed imaging data on these malformations as well as surgical considerations and outcomes.
Conflict of interest statement
The authors have no financial interests or disclosures to declare.
Figures

References
-
- Potti T. A., Petty E. M., Lesperance M. M. A comprehensive review of reported heritable noggin-associated syndromes and proposed clinical utility of one broadly inclusive diagnostic term: NOG-related-symphalangism spectrum disorder (NOG-SSD) Human Mutation. 2011;32(8):877–886. doi: 10.1002/humu.21515. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

