Development and validation of an interferon signature predicting prognosis and treatment response for glioblastoma
- PMID: 31428519
- PMCID: PMC6685507
- DOI: 10.1080/2162402X.2019.1621677
Development and validation of an interferon signature predicting prognosis and treatment response for glioblastoma
Abstract
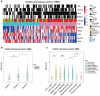
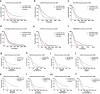
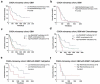
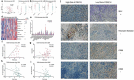
Background: Interferon treatment, as an important approach of anti-tumor immunotherapy, has been implemented in multiple clinical trials of glioma. However, only a small number of gliomas benefit from it. Therefore, it is necessary to investigate the clinical role of interferons and to establish robust biomarkers to facilitate its application. Materials and methods: This study reviewed 1,241 glioblastoma (GBM) and 1,068 lower grade glioma (LGG) patients from six glioma cohorts. The transcription matrix and clinical information were analyzed using R software, GraphPad Prism 7 and Medcalc, etc. Immunohistochemical (IHC) staining were performed for validation in protein level. Results: Interferon signaling was significantly enhanced in GBM. An interferon signature was developed based on five interferon genes with prognostic significance, which could reflect various interferon statuses. Survival analysis showed the signature could serve as an unfavorable prognostic factor independently. We also established a nomogram model integrating the risk signature into traditional prognostic factors, which increased the validity of survival prediction. Moreover, high-risk group conferred resistance to chemotherapy and high IFNB1 expression levels. Functional analysis showed that the high-risk group was associated with overloaded immune response. Microenvironment analysis and IHC staining found that high-risk group occupied a disorganized microenvironment which was characterized by an enrichment of M0 macrophages and neutrophils, but less infiltration of activated nature killing (NK) cells and M1 type macrophages. Conclusion: This interferon signature was an independent indicator for unfavorable prognosis and showed great potential for screening out patients who will benefit from chemotherapy and interferon treatment.
Keywords: Glioblastoma; immune response; interferon; microenvironment; prognosis.
Figures

Similar articles
-
A novel defined risk signature of interferon response genes predicts the prognosis and correlates with immune infiltration in glioblastoma.Math Biosci Eng. 2022 Jun 29;19(9):9481-9504. doi: 10.3934/mbe.2022441. Math Biosci Eng. 2022. PMID: 35942769
-
Characterization of Hypoxia Signature to Evaluate the Tumor Immune Microenvironment and Predict Prognosis in Glioma Groups.Front Oncol. 2020 May 15;10:796. doi: 10.3389/fonc.2020.00796. eCollection 2020. Front Oncol. 2020. PMID: 32500034 Free PMC article.
-
A Risk Classification System With Five-Gene for Survival Prediction of Glioblastoma Patients.Front Neurol. 2019 Jul 16;10:745. doi: 10.3389/fneur.2019.00745. eCollection 2019. Front Neurol. 2019. PMID: 31379707 Free PMC article.
-
Identification of potential biomarkers related to glioma survival by gene expression profile analysis.BMC Med Genomics. 2019 Mar 20;11(Suppl 7):34. doi: 10.1186/s12920-019-0479-6. BMC Med Genomics. 2019. PMID: 30894197 Free PMC article.
-
Bioinformatic profiling identifies an immune-related risk signature for glioblastoma.Neurology. 2016 Jun 14;86(24):2226-34. doi: 10.1212/WNL.0000000000002770. Epub 2016 May 25. Neurology. 2016. PMID: 27225222
Cited by
-
Promoting Prognostic Model Application: A Review Based on Gliomas.J Oncol. 2021 Jul 31;2021:7840007. doi: 10.1155/2021/7840007. eCollection 2021. J Oncol. 2021. PMID: 34394352 Free PMC article. Review.
-
Inhibiting interferon-γ induced cancer intrinsic TNFRSF14 elevation restrains the malignant progression of glioblastoma.J Exp Clin Cancer Res. 2024 Jul 31;43(1):212. doi: 10.1186/s13046-024-03131-7. J Exp Clin Cancer Res. 2024. PMID: 39085878 Free PMC article.
-
Recognition of Differentially Expressed Molecular Signatures and Pathways Associated with COVID-19 Poor Prognosis in Glioblastoma Patients.Int J Mol Sci. 2023 Feb 10;24(4):3562. doi: 10.3390/ijms24043562. Int J Mol Sci. 2023. PMID: 36834974 Free PMC article.
-
A nomogram-based immune-serum scoring system predicts overall survival in patients with lung adenocarcinoma.Cancer Biol Med. 2021 Mar 12;18(2):517-29. doi: 10.20892/j.issn.2095-3941.2020.0648. Online ahead of print. Cancer Biol Med. 2021. PMID: 33710816 Free PMC article.
-
NLRC5: A Potential Target for Central Nervous System Disorders.Front Immunol. 2021 Jun 18;12:704989. doi: 10.3389/fimmu.2021.704989. eCollection 2021. Front Immunol. 2021. PMID: 34220868 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
