Beyond the Trinity of ATM, ATR, and DNA-PK: Multiple Kinases Shape the DNA Damage Response in Concert With RNA Metabolism
- PMID: 31428617
- PMCID: PMC6688092
- DOI: 10.3389/fmolb.2019.00061
Beyond the Trinity of ATM, ATR, and DNA-PK: Multiple Kinases Shape the DNA Damage Response in Concert With RNA Metabolism
Abstract
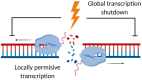
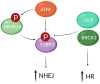
Our genome is constantly exposed to endogenous and exogenous sources of DNA damage resulting in various alterations of the genetic code. DNA double-strand breaks (DSBs) are considered one of the most cytotoxic lesions. Several types of repair pathways act to repair DNA damage and maintain genome stability. In the canonical DNA damage response (DDR) DSBs are recognized by the sensing kinases Ataxia-telangiectasia mutated (ATM), Ataxia-telangiectasia and Rad3-related (ATR), and DNA-dependent protein kinase (DNA-PK), which initiate a cascade of kinase-dependent amplification steps known as DSB signaling. Recent evidence suggests that efficient recognition and repair of DSBs relies on the transcription and processing of non-coding (nc)RNA molecules by RNA polymerase II (RNAPII) and the RNA interference (RNAi) factors Drosha and Dicer. Multiple kinases influence the phosphorylation status of both the RNAPII carboxy-terminal domain (CTD) and Dicer in order to regulate RNA-dependent DSBs repair. The importance of kinase signaling and RNA processing in the DDR is highlighted by the regulation of p53-binding protein (53BP1), a key regulator of DSB repair pathway choice between homologous recombination (HR) and non-homologous end joining (NHEJ). Additionally, emerging evidence suggests that RNA metabolic enzymes also play a role in the repair of other types of DNA damage, including the DDR to ultraviolet radiation (UVR). RNAi factors are also substrates for mitogen-activated protein kinase (MAPK) signaling and mediate the turnover of ncRNA during nucleotide excision repair (NER) in response to UVR. Here, we review kinase-dependent phosphorylation events on RNAPII, Drosha and Dicer, and 53BP1 that modulate the key steps of the DDR to DSBs and UVR, suggesting an intimate link between the DDR and RNA metabolism.
Keywords: 53BP1; DNA damage response; RNA metabolism; RNA polymerase II; dicer; kinase; phosphorylation.
Figures
References
-
- Altafaj X., Dierssen M., Baamonde C., Marti E., Visa J., Guimera J., et al. . (2001). Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum. Mol. Genet. 10, 1915–1923. 10.1093/hmg/10.18.1915 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

