Homologous recombination DNA repair defects in PALB2- associated breast cancers
- PMID: 31428676
- PMCID: PMC6687719
- DOI: 10.1038/s41523-019-0115-9
Homologous recombination DNA repair defects in PALB2- associated breast cancers
Erratum in
-
Erratum: Publisher Correction: Homologous recombination DNA repair defects in PALB2-associated breast cancers.NPJ Breast Cancer. 2019 Nov 19;5:44. doi: 10.1038/s41523-019-0140-8. eCollection 2019. NPJ Breast Cancer. 2019. PMID: 31754629 Free PMC article.
Abstract
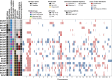
Mono-allelic germline pathogenic variants in the Partner And Localizer of BRCA2 (PALB2) gene predispose to a high-risk of breast cancer development, consistent with the role of PALB2 in homologous recombination (HR) DNA repair. Here, we sought to define the repertoire of somatic genetic alterations in PALB2-associated breast cancers (BCs), and whether PALB2-associated BCs display bi-allelic inactivation of PALB2 and/or genomic features of HR-deficiency (HRD). Twenty-four breast cancer patients with pathogenic PALB2 germline mutations were analyzed by whole-exome sequencing (WES, n = 16) or targeted capture massively parallel sequencing (410 cancer genes, n = 8). Somatic genetic alterations, loss of heterozygosity (LOH) of the PALB2 wild-type allele, large-scale state transitions (LSTs) and mutational signatures were defined. PALB2-associated BCs were found to be heterogeneous at the genetic level, with PIK3CA (29%), PALB2 (21%), TP53 (21%), and NOTCH3 (17%) being the genes most frequently affected by somatic mutations. Bi-allelic PALB2 inactivation was found in 16 of the 24 cases (67%), either through LOH (n = 11) or second somatic mutations (n = 5) of the wild-type allele. High LST scores were found in all 12 PALB2-associated BCs with bi-allelic PALB2 inactivation sequenced by WES, of which eight displayed the HRD-related mutational signature 3. In addition, bi-allelic inactivation of PALB2 was significantly associated with high LST scores. Our findings suggest that the identification of bi-allelic PALB2 inactivation in PALB2-associated BCs is required for the personalization of HR-directed therapies, such as platinum salts and/or PARP inhibitors, as the vast majority of PALB2-associated BCs without PALB2 bi-allelic inactivation lack genomic features of HRD.
Keywords: Breast cancer; Cancer genetics; Cancer genomics.
Conflict of interest statement
Competing interestsM.E.R. reports consultancy fees from McKesson and AstraZeneca, and uncompensated consulting/advisory activities with Merck and Pfizer. J.S.R.-F. reports personal/consultancy fees from VolitionRx, Page.AI, Goldman Sachs, Grail, Ventana Medical Systems, Invicro, and Genentech, outside the scope of the submitted work. All remaining authors declare no competing interests.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

