RhoBTB1 interacts with ROCKs and inhibits invasion
- PMID: 31431478
- PMCID: PMC6744581
- DOI: 10.1042/BCJ20190203
RhoBTB1 interacts with ROCKs and inhibits invasion
Abstract
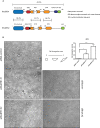
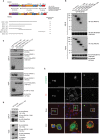
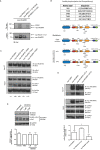
RhoBTB1 is an atypical Rho GTPase with two BTB domains in addition to its Rho domain. Although most Rho GTPases regulate actin cytoskeletal dynamics, RhoBTB1 is not known to affect cell shape or motility. We report that RhoBTB1 depletion increases prostate cancer cell invasion and induces elongation in Matrigel, a phenotype similar to that induced by depletion of ROCK1 and ROCK2. We demonstrate that RhoBTB1 associates with ROCK1 and ROCK2 and its association with ROCK1 is via its Rho domain. The Rho domain binds to the coiled-coil region of ROCK1 close to its kinase domain. We identify two amino acids within the Rho domain that alter RhoBTB1 association with ROCK1. RhoBTB1 is a substrate for ROCK1, and mutation of putative phosphorylation sites reduces its association with Cullin3, a scaffold for ubiquitin ligases. We propose that RhoBTB1 suppresses cancer cell invasion through interacting with ROCKs, which in turn regulate its association with Cullin3. Via Cullin3, RhoBTB1 has the potential to affect protein degradation.
Keywords: Cullin3; Rho GTPases; Rho-kinases; RhoBTB; cell invasion; phosphorylation.
© 2019 The Author(s).
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures



Similar articles
-
Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes--evidence for an autoregulatory mechanism.Exp Cell Res. 2008 Nov 15;314(19):3453-65. doi: 10.1016/j.yexcr.2008.09.005. Epub 2008 Sep 20. Exp Cell Res. 2008. PMID: 18835386 Free PMC article.
-
Rho GTPases of the RhoBTB subfamily and tumorigenesis.Acta Pharmacol Sin. 2008 Mar;29(3):285-95. doi: 10.1111/j.1745-7254.2008.00773.x. Acta Pharmacol Sin. 2008. PMID: 18298893 Review.
-
RhoBTB1 protects against hypertension and arterial stiffness by restraining phosphodiesterase 5 activity.J Clin Invest. 2019 Mar 21;129(6):2318-2332. doi: 10.1172/JCI123462. J Clin Invest. 2019. PMID: 30896450 Free PMC article.
-
The tumor suppressor RhoBTB1 controls Golgi integrity and breast cancer cell invasion through METTL7B.BMC Cancer. 2017 Feb 20;17(1):145. doi: 10.1186/s12885-017-3138-3. BMC Cancer. 2017. PMID: 28219369 Free PMC article.
-
Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization.Biochim Biophys Acta Gen Subj. 2017 May;1861(5 Pt A):1009-1023. doi: 10.1016/j.bbagen.2017.02.005. Epub 2017 Feb 8. Biochim Biophys Acta Gen Subj. 2017. PMID: 28188861 Review.
Cited by
-
Emerging roles for Rho GTPases operating at the Golgi complex.Small GTPases. 2021 Sep-Nov;12(5-6):311-322. doi: 10.1080/21541248.2020.1812873. Epub 2020 Sep 3. Small GTPases. 2021. PMID: 32881632 Free PMC article. Review.
-
Cullin-3: Renal and Vascular Mechanisms Regulating Blood Pressure.Curr Hypertens Rep. 2020 Aug 27;22(9):61. doi: 10.1007/s11906-020-01076-8. Curr Hypertens Rep. 2020. PMID: 32852625 Free PMC article. Review.
-
The metastasis-promoting P1597L mutation in PlexinB1 enhances Ras activity.BMC Cancer. 2024 Aug 13;24(1):1004. doi: 10.1186/s12885-024-12762-0. BMC Cancer. 2024. PMID: 39138404 Free PMC article.
-
Unravelling cell migration: defining movement from the cell surface.Cell Adh Migr. 2022 Dec;16(1):25-64. doi: 10.1080/19336918.2022.2055520. Cell Adh Migr. 2022. PMID: 35499121 Free PMC article. Review.
-
RhoBTB1 reverses established arterial stiffness in angiotensin II-induced hypertension by promoting actin depolymerization.JCI Insight. 2022 May 9;7(9):e158043. doi: 10.1172/jci.insight.158043. JCI Insight. 2022. PMID: 35358093 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

