Myeloid and CD4 T Cells Comprise the Latent Reservoir in Antiretroviral Therapy-Suppressed SIVmac251-Infected Macaques
- PMID: 31431552
- PMCID: PMC6703426
- DOI: 10.1128/mBio.01659-19
Myeloid and CD4 T Cells Comprise the Latent Reservoir in Antiretroviral Therapy-Suppressed SIVmac251-Infected Macaques
Abstract
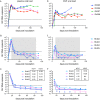
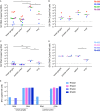
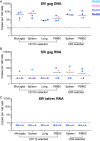
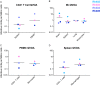
Human immunodeficiency virus (HIV) eradication or long-term suppression in the absence of antiretroviral therapy (ART) requires an understanding of all viral reservoirs that could contribute to viral rebound after ART interruption. CD4 T cells (CD4s) are recognized as the predominant reservoir in HIV type 1 (HIV-1)-infected individuals. However, macrophages are also infected by HIV-1 and simian immunodeficiency virus (SIV) during acute infection and may persist throughout ART, contributing to the size of the latent reservoir. We sought to determine whether tissue macrophages contribute to the SIVmac251 reservoir in suppressed macaques. Using cell-specific quantitative viral outgrowth assays (CD4-QVOA and MΦ-QVOA), we measured functional latent reservoirs in CD4s and macrophages in ART-suppressed SIVmac251-infected macaques. Spleen, lung, and brain in all suppressed animals contained latently infected macrophages, undetectable or low-level SIV RNA, and detectable SIV DNA. Silent viral genomes with potential for reactivation and viral spread were also identified in blood monocytes, although these cells might not be considered reservoirs due to their short life span. Additionally, virus produced in the MΦ-QVOA was capable of infecting healthy activated CD4s. Our results strongly suggest that functional latent reservoirs in CD4s and macrophages can contribute to viral rebound and reestablishment of productive infection after ART interruption. These findings should be considered in the design and implementation of future HIV cure strategies.IMPORTANCE This study provides further evidence that the latent reservoir is comprised of both CD4+ T cells and myeloid cells. The data presented here suggest that CD4+ T cells and macrophages found throughout tissues in the body can contain replication-competent SIV and contribute to rebound of the virus after treatment interruption. Additionally, we have shown that monocytes in blood contain latent virus and, though not considered a reservoir themselves due to their short life span, could contribute to the size of the latent reservoir upon entering the tissue and differentiating into long-lived macrophages. These new insights into the size and location of the SIV reservoir using a model that is heavily studied in the HIV field could have great implications for HIV-infected individuals and should be taken into consideration with the development of future HIV cure strategies.
Keywords: HIV; SIV; latency; macrophages; monocytes.
Copyright © 2019 Abreu et al.
Figures

References
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi: 10.1038/8394. - DOI - PubMed
-
- Avalos CR, Abreu CM, Queen SE, Li M, Price S, Shirk EN, Engle EL, Forsyth E, Bullock BT, Mac Gabhann F, Wietgrefe SW, Haase AT, Zink MC, Mankowski JL, Clements JE, Gama L. 2017. Brain macrophages in simian immunodeficiency virus-infected, antiretroviral-suppressed macaques: a functional latent reservoir. mBio 8:e01186-17. doi: 10.1128/mBio.01186-17. - DOI - PMC - PubMed
-
- Avalos CR, Price SL, Forsyth ER, Pin JN, Shirk EN, Bullock BT, Queen SE, Li M, Gellerup D, O’Connor SL, Zink MC, Mankowski JL, Gama L, Clements JE. 2016. Quantitation of productively infected monocytes and macrophages of simian immunodeficiency virus-infected macaques. J Virol 90:5643–5656. doi: 10.1128/JVI.00290-16. - DOI - PMC - PubMed
-
- Abreu CM, Veenhuis RT, Avalos CR, Graham S, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Clements JE, Gama L. 2019. Infectious virus persists in CD4+ T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J Virol 93:e00065-19. doi: 10.1128/JVI.00065-19. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

