The Δ133p53β isoform promotes an immunosuppressive environment leading to aggressive prostate cancer
- PMID: 31431617
- PMCID: PMC6702175
- DOI: 10.1038/s41419-019-1861-1
The Δ133p53β isoform promotes an immunosuppressive environment leading to aggressive prostate cancer
Abstract
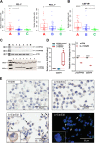
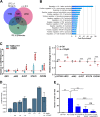
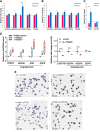
Prostate cancer is the second most common cancer in men, for which there are no reliable biomarkers or targeted therapies. Here we demonstrate that elevated levels of Δ133TP53β isoform characterize prostate cancers with immune cell infiltration, particularly T cells and CD163+ macrophages. These cancers are associated with shorter progression-free survival, Gleason scores ≥ 7, and an immunosuppressive environment defined by a higher proportion of PD-1, PD-L1 and colony-stimulating factor 1 receptor (CSF1R) positive cells. Consistent with this, RNA-seq of tumours showed enrichment for pathways associated with immune signalling and cell migration. We further show a role for hypoxia and wild-type p53 in upregulating Δ133TP53 levels. Finally, AUC analysis showed that Δ133TP53β expression level alone predicted aggressive disease with 88% accuracy. Our data identify Δ133TP53β as a highly accurate prognostic factor for aggressive prostate cancer.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Ecke TH, et al. TP53 gene mutations in prostate cancer progression. Anticancer Res. 2010;30:1579–1586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

