Hybridization drives genetic erosion in sympatric desert fishes of western North America
- PMID: 31431737
- PMCID: PMC6834602
- DOI: 10.1038/s41437-019-0259-2
Hybridization drives genetic erosion in sympatric desert fishes of western North America
Abstract
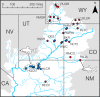
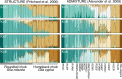
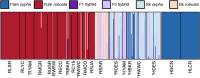
Many species have evolved or currently coexist in sympatry due to differential adaptation in a heterogeneous environment. However, anthropogenic habitat modifications can either disrupt reproductive barriers or obscure environmental conditions which underlie fitness gradients. In this study, we evaluated the potential for an anthropogenically-mediated shift in reproductive boundaries that separate two historically sympatric fish species (Gila cypha and G. robusta) endemic to the Colorado River Basin using ddRAD sequencing of 368 individuals. We first examined the integrity of reproductive isolation while in sympatry and allopatry, then characterized hybrid ancestries using genealogical assignment tests. We tested for localized erosion of reproductive isolation by comparing site-wise genomic clines against global patterns and identified a breakdown in the drainage-wide pattern of selection against interspecific heterozygotes. This, in turn, allowed for the formation of a hybrid swarm in one tributary, and asymmetric introgression where species co-occur. We also detected a weak but significant relationship between genetic purity and degree of consumptive water removal, suggesting a role for anthropogenic habitat modifications in undermining species boundaries or expanding historically limited introgression. In addition, results from basin-wide genomic clines suggested that hybrids and parental forms are adaptively nonequivalent. If so, then a failure to manage for hybridization will exacerbate the long-term extinction risk in parental populations. These results reinforce the role of anthropogenic habitat modification in promoting interspecific introgression in sympatric species by relaxing divergent selection. This, in turn, underscores a broader role for hybridization in decreasing global biodiversity within rapidly deteriorating environments.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

