USP33 deubiquitinates PRKN/parkin and antagonizes its role in mitophagy
- PMID: 31432739
- PMCID: PMC7138199
- DOI: 10.1080/15548627.2019.1656957
USP33 deubiquitinates PRKN/parkin and antagonizes its role in mitophagy
Abstract
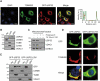
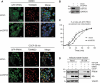
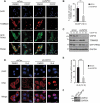
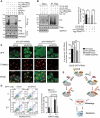
PRKN/parkin activation through phosphorylation of its ubiquitin and ubiquitin-like domain by PINK1 is critical in mitophagy induction for eliminating the damaged mitochondria. Deubiquitinating enzymes (DUBs) functionally reversing PRKN ubiquitination are critical in controlling the magnitude of PRKN-mediated mitophagy process. However, potential DUBs that directly target PRKN and antagonize its pro-mitophagy effect remains to be identified and characterized. Here, we demonstrated that USP33/VDU1 is localized at the outer membrane of mitochondria and serves as a PRKN DUB through their interaction. Cellular and in vitro assays illustrated that USP33 deubiquitinates PRKN in a DUB activity-dependent manner. USP33 prefers to remove K6, K11, K48 and K63-linked ubiquitin conjugates from PRKN, and deubiquitinates PRKN mainly at Lys435. Mutation of this site leads to a significantly decreased level of K63-, but not K48-linked PRKN ubiquitination. USP33 deficiency enhanced both K48- and K63-linked PRKN ubiquitination, but only K63-linked PRKN ubiquitination was significantly increased under mitochondrial depolarization. Further, USP33 knockdown increased both PRKN protein stabilization and its translocation to depolarized mitochondria leading to the enhancement of mitophagy. Moreover, USP33 silencing protects SH-SY5Y human neuroblastoma cells from the neurotoxin MPTP-induced apoptotic cell death. Our findings convincingly demonstrate that USP33 is a novel PRKN deubiquitinase antagonizing its regulatory roles in mitophagy and SH-SY5Y neuron-like cell survival. Thus, USP33 inhibition may represents an attractive new therapeutic strategy for PD patients.Abbreviations: CCCP: carbonyl cyanide 3-chlorophenylhydrazone; DUB: deubiquitinating enzymes; MPTP: 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; OMM: outer mitochondrial membrane; PD: Parkinson disease; PINK1: PTEN induced kinase 1; PRKN/PARK2: parkin RBR E3 ubiquitin protein ligase; ROS: reactive oxygen species; TM: transmembrane; Ub: ubiquitin; UBA1: ubiquitin like modifier activating enzyme 1; UBE2L3/UbcH7: ubiquitin conjugating enzyme E2 L3; USP33: ubiquitin specific peptidase 33; WT: wild type.
Keywords: Apoptosis; PRKN/parkin; USP33 deubiquitinase; mitophagy; ubiquitination.
Figures


References
-
- Bingol B, Tea JS, Phu L, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510(7505):370–375. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
