Strong anion exchange-mediated phosphoproteomics reveals extensive human non-canonical phosphorylation
- PMID: 31433507
- PMCID: PMC6826212
- DOI: 10.15252/embj.2018100847
Strong anion exchange-mediated phosphoproteomics reveals extensive human non-canonical phosphorylation
Abstract
Phosphorylation is a key regulator of protein function under (patho)physiological conditions, and defining site-specific phosphorylation is essential to understand basic and disease biology. In vertebrates, the investigative focus has primarily been on serine, threonine and tyrosine phosphorylation, but mounting evidence suggests that phosphorylation of other "non-canonical" amino acids also regulates critical aspects of cell biology. However, standard methods of phosphoprotein characterisation are largely unsuitable for the analysis of non-canonical phosphorylation due to their relative instability under acidic conditions and/or elevated temperature. Consequently, the complete landscape of phosphorylation remains unexplored. Here, we report an unbiased phosphopeptide enrichment strategy based on strong anion exchange (SAX) chromatography (UPAX), which permits identification of histidine (His), arginine (Arg), lysine (Lys), aspartate (Asp), glutamate (Glu) and cysteine (Cys) phosphorylation sites on human proteins by mass spectrometry-based phosphoproteomics. Remarkably, under basal conditions, and having accounted for false site localisation probabilities, the number of unique non-canonical phosphosites is approximately one-third of the number of observed canonical phosphosites. Our resource reveals the previously unappreciated diversity of protein phosphorylation in human cells, and opens up avenues for high-throughput exploration of non-canonical phosphorylation in all organisms.
Keywords: mass spectrometry; non-canonical phosphorylation; phosphohistidine; phosphoproteomics; strong anion exchange chromatography.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
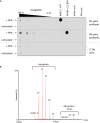
PPA‐treated myoglobin preferentially generates N3‐phosphohistidine. Serial dilutions of myoglobin and PPA‐treated myoglobin were dotted onto nitrocellulose membrane and incubated with either the N1‐ or N3‐phosphohistidine antibody (or secondary antibody only), as indicated. Phosphorylated PGAM and NME1 proteins were also dotted onto the membrane, along with their unphosphorylated forms, as controls for the N1‐ and N3‐pHis antibodies, respectively. PGAM: phosphoglycerate mutase. NME1: nucleoside diphosphate kinase.
Zero charge state mass spectrum of intact phosphorylated myoglobin. Phosphorylated myoglobin was analysed by direct infusion via nano‐ESI into a Synapt G2‐Si mass spectrometer. The raw mass spectrum was deconvoluted using MaxEnt1. Up to five phosphate groups are observed per intact myoglobin molecule. Also apparent is the haem‐bound form of myoglobin containing up to four phosphate groups.
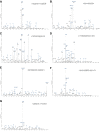
- A–G
The identified phosphorylation site is indicated (*), and the sequence is detailed on the mass spectrum. (A) doubly charged ion at m/z 843.9: pHis25; (B) doubly charged ion at m/z 676.3: pHis37; (C) doubly charged ion at m/z 729.9: pHis65; (D) triply charged ion at m/z 645.3: pHis82; (E) ETD spectrum of triply charged ion at m/z 645.3: pHis83; (F) triply charged ion at m/z 645.3: pHis94; and (G) triply charged ion at m/z 655.6: pHis114. The triplet neutral loss ions can be observed in all six HCD spectra (A–D, F–G) for pHis‐containing peptides from myoglobin.
- A–C
Percentage of the total signal intensity attributed to phosphopeptides (blue bars, 11 unique peptide ions) and non‐phosphorylated peptides (grey bars, 25 unique peptide ions) following SAX fractionation at (A) pH 6.0, (B) pH 6.8 and (C) pH 8.0, with later fractions consisting of up to 100% phosphopeptide ion signal.
- D
Representative pHis‐containing myoglobin peptide (LFTG
H PETLEK, solid line) and its non‐phosphorylated counterpart (LFTGHPETLEK, dashed line), quantified across all 16 SAX fractions; SAX was performed at pH 6.0 (red), pH 6.8 (blue) or pH 8.0 (green). Percentage of the total peak area of each individual peptide across the gradient is plotted for each fraction. In order to assess pHis stability and effects of SAX separation in a complex mixture (black), SAX was also repeated at pH 6.8 with phosphorylated myoglobin spiked into a human cell lysate prior to digestion. Data for the five pHis‐containing myoglobin peptides are shown in Fig EV3.

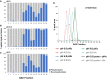
- A
Schematic representation of the UPAX strategy for phosphopeptide enrichment and MS/MS analyses.
- B
siRNA‐mediated knockdown of PHPT1 in HeLa cells (24 h) with reference to GAPDH loading control. NT—non‐targeting siRNA control.
- C–F
(C) Representative SAX profile of trypsin‐digested HeLa lysate (Abs280 nm). Base peak chromatograms are shown for select SAX fractions following high‐resolution LC‐MS/MS using an Orbitrap Fusion mass spectrometer: (D) fraction 3 (green); (E) fraction 6 (red); and (F) fraction 10 (grey). Peptides were fragmented by HCD, with neutral loss of 98 amu from the precursor ions triggering EThcD (Ferries et al, 2017). Tandem mass spectra were separated according to fragmentation strategy in Proteome Discoverer prior to searching with Mascot. The ptmRS node was used for phosphosite localisation. Analysis was performed on three independent biological replicates for each condition (NT or PHPT1 siRNA).
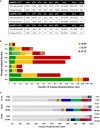
Total number of phosphopeptides identified (5% FDR) and unique sites for each phosphorylated residue according to site localisation confidence (ptmRS score). For pLys and pArg, the number in parentheses is the total number of identified sites/peptides including those localised to the peptide C‐terminus; outside of parentheses are the non‐C‐terminally mapped pLys or pArg sites.
Number of unique phosphorylation sites defined at different site localisation confidence values: ptmRS ≥ 0.99 (green); ptmRS ≥ 0.90 (yellow); and ptmRS ≥ 0.75 (red). Light green, yellow or red indicates those sites of pLys or pArg mapped to the extreme peptide C‐terminal residue at each ptmRS score cut‐off.
Percentage of canonical pSer, pThr and pTyr sites (shades of grey) compared with non‐canonical phosphosites (pHis—yellow; pAsp—blue; pGlu—green; pLys—red; pArg—purple) identified using either the described UPAX strategy or a standard TiO2‐based phosphopeptide enrichment protocol, as a function of ptmRS score.
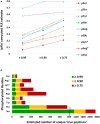
pAla‐computed FLR estimate for each type of canonical and non‐canonical phosphorylated residue at ptmRS values ≥ 0.99, ≥ 0.90 or ≥ 0.75.
Total number of phosphorylation sites estimated to be “true positives” based on the pAla‐determined residue‐specific FLR, for each phosphorylated residue according to ptmRS score. pLys* and pArg* represent non‐C‐terminal pLys or pArg residues, respectively (see Appendix Supplementary Methods).
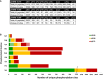
For pLys and pArg, the number in parentheses is the total number of identified sites/peptides including those localised to the peptide C‐terminus. Non‐C‐terminally mapped pLys or pArg sites are outside of the parentheses.
The chart displays the total number of unique phosphorylation sites defined at different site localisation confidence values: ptmRS ≥ 0.99 (green); ptmRS ≥ 0.90 (yellow); and ptmRS ≥ 0.75 (red). Light green, yellow or red indicates those sites of pLys or pArg mapped to the extreme peptide C‐terminal residue at each ptmRS score cut‐off.
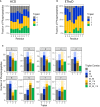
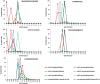
Comparison of triplet score (≥ 5% S/N) following HCD of unique phosphopeptides (5% FDR) identified from UPAX separated tryptic digests of HeLa cell‐derived phosphopeptides. Site of phosphorylation (ptmRS score ≥ 0.90) is indicated. Percentage of phosphopeptides exhibiting either no neutral loss (triplet = 0) or neutral loss of any 1 (triplet = 1), 2 (triplet = 2) or 3 ions from the precursor (∆80, ∆98 and/or ∆116 amu).
Distribution of triplet score following HCD fragmentation as a function of site localisation confidence (ptmRS score ≥ 0.75, ≥ 0.90, ≥ 0.95 and ≥ 0.99) for unique singly phosphorylated peptide as a function of the residue phosphorylated.
Numbers of phosphopeptides (ptmRS ≥ 0.90) exhibiting different combinations of the three neutral loss species (∆80, ∆98 and ∆116 amu from precursor) for each of the phosphorylated residues.
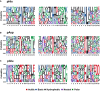
- A–C
The amino acid sequences surrounding confidently localised sites of (A) pHis, (B) pAsp and (C) pGlu (ptmRS ≥ 0.99) were analysed for sequence enrichment using Motif‐X. Depicted are the sequences of the enriched motifs. Additional details are presented in Appendix Figs S8, S11 and S13.

- A, B
Functional annotation of the significantly enriched (A) pAsp‐containing proteins or (B) pGlu‐containing proteins (ptmRS ≥ 0.90) using DAVID (q‐value < 0.05). BH: Benjamini–Hochberg.

- A–C
The amino acid sequences surrounding confidently localised sites of (A) non‐C‐terminally localised pLys, (B) non‐C‐terminally localised pArg and (C) pCys (ptmRS ≥ 0.99) were analysed for sequence enrichment using Motif‐X. Depicted are the sequences of the enriched motifs. Additional details are presented in Appendix Figs S15, S17 and S18.
References
-
- Attwood PV, Piggott MJ, Zu XL, Besant PG (2007) Focus on phosphohistidine. Amino Acids 32: 145–156 - PubMed
-
- Attwood PV, Besant PG, Piggott MJ (2011) Focus on phosphoaspartate and phosphoglutamate. Amino Acids 40: 1035–1051 - PubMed
-
- Attwood PV (2013) Histidine kinases from bacteria to humans. Biochem Soc Trans 41: 1023–1028 - PubMed
-
- Bentley‐DeSousa A, Downey M (2019) From underlying chemistry to therapeutic potential: open questions in the new field of lysine polyphosphorylation. Curr Genet 65: 57–64 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/M023818/1/UK Research and Innovation|Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/L005239/1/UK Research and Innovation|Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/N021703/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H007113/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M012557/1/UK Research and Innovation|Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/R02216X/1/UK Research and Innovation|Biotechnology and Biological Sciences Research Council (BBSRC)/International
- CR1157/North West Cancer Research Fund (NWCR)/International
- BB/H007113/1/UK Research and Innovation|Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/M025705/1/UK Research and Innovation|Biotechnology and Biological Sciences Research Council (BBSRC)/International
- CR1088/North West Cancer Research Fund (NWCR)/International
- CR1037/North West Cancer Research Fund (NWCR)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

