Transient crosslinking kinetics optimize gene cluster interactions
- PMID: 31433796
- PMCID: PMC6730938
- DOI: 10.1371/journal.pcbi.1007124
Transient crosslinking kinetics optimize gene cluster interactions
Abstract
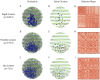
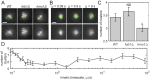
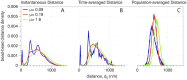
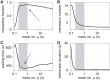
Our understanding of how chromosomes structurally organize and dynamically interact has been revolutionized through the lens of long-chain polymer physics. Major protein contributors to chromosome structure and dynamics are condensin and cohesin that stochastically generate loops within and between chains, and entrap proximal strands of sister chromatids. In this paper, we explore the ability of transient, protein-mediated, gene-gene crosslinks to induce clusters of genes, thereby dynamic architecture, within the highly repeated ribosomal DNA that comprises the nucleolus of budding yeast. We implement three approaches: live cell microscopy; computational modeling of the full genome during G1 in budding yeast, exploring four decades of timescales for transient crosslinks between 5kbp domains (genes) in the nucleolus on Chromosome XII; and, temporal network models with automated community (cluster) detection algorithms applied to the full range of 4D modeling datasets. The data analysis tools detect and track gene clusters, their size, number, persistence time, and their plasticity (deformation). Of biological significance, our analysis reveals an optimal mean crosslink lifetime that promotes pairwise and cluster gene interactions through "flexible" clustering. In this state, large gene clusters self-assemble yet frequently interact (merge and separate), marked by gene exchanges between clusters, which in turn maximizes global gene interactions in the nucleolus. This regime stands between two limiting cases each with far less global gene interactions: with shorter crosslink lifetimes, "rigid" clustering emerges with clusters that interact infrequently; with longer crosslink lifetimes, there is a dissolution of clusters. These observations are compared with imaging experiments on a normal yeast strain and two condensin-modified mutant cell strains. We apply the same image analysis pipeline to the experimental and simulated datasets, providing support for the modeling predictions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Marko JF, Siggia ED. Fluctuations and supercoiling of DNA. Science. 1994;265(5171):506–508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

