Distinct serum biosignatures are associated with different tuberculosis treatment outcomes
- PMID: 31434026
- PMCID: PMC6839616
- DOI: 10.1016/j.tube.2019.101859
Distinct serum biosignatures are associated with different tuberculosis treatment outcomes
Abstract
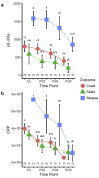
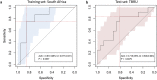
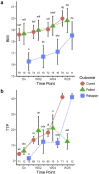
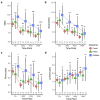
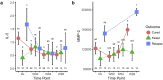
Biomarkers for TB treatment response and outcome are needed. This study characterize changes in immune profiles during TB treatment, define biosignatures associated with treatment outcomes, and explore the feasibility of predictive models for relapse. Seventy-two markers were measured by multiplex cytokine array in serum samples from 78 cured, 12 relapsed and 15 failed treatment patients from South Africa before and during therapy for pulmonary TB. Promising biosignatures were evaluated in a second cohort from Uganda/Brazil consisting of 17 relapse and 23 cured patients. Thirty markers changed significantly with different response patterns during TB treatment in cured patients. The serum biosignature distinguished cured from relapse patients and a combination of two clinical (time to positivity in liquid culture and BMI) and four immunological parameters (TNF-β, sIL-6R, IL-12p40 and IP-10) at diagnosis predicted relapse with a 75% sensitivity (95%CI 0.38-1) and 85% specificity (95%CI 0.75-0.93). This biosignature was validated in an independent Uganda/Brazil cohort correctly classifying relapse patients with 83% (95%CI 0.58-1) sensitivity and 61% (95%CI 0.39-0.83) specificity. A characteristic biosignature with value as predictor of TB relapse was identified. The repeatability and robustness of these biomarkers require further validation in well-characterized cohorts.
Keywords: Biomarkers; Relapse; Treatment failure; Tuberculosis; Tuberculosis treatment.
Copyright © 2019 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




References
-
- Hong Kong Chest Service/Tuberculosis Research Center Madras/British Medical Research C. A controlled trial of a 2-month, 3-month, and 12-month regimens of chemotherapy for sputum smear-negative pulmonary tuberculosis: the results at 30 months. Hong Kong Chest Service/Tuberculosis Research Centre, Madras/British Medical Research Council. Am Rev Respir Dis. 1981;124(2):138–142. - PubMed
-
- Mitchison D.A. Assessment of new sterilizing drugs for treating pulmonary tuberculosis by culture at 2 months. Am Rev Respir Dis. 1993;147(4):1062–1063. - PubMed
-
- Wallis R.S. Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect Dis. 2010;10(2):68–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

