Chromosomics: Bridging the Gap between Genomes and Chromosomes
- PMID: 31434289
- PMCID: PMC6723020
- DOI: 10.3390/genes10080627
Chromosomics: Bridging the Gap between Genomes and Chromosomes
Abstract
The recent advances in DNA sequencing technology are enabling a rapid increase in the number of genomes being sequenced. However, many fundamental questions in genome biology remain unanswered, because sequence data alone is unable to provide insight into how the genome is organised into chromosomes, the position and interaction of those chromosomes in the cell, and how chromosomes and their interactions with each other change in response to environmental stimuli or over time. The intimate relationship between DNA sequence and chromosome structure and function highlights the need to integrate genomic and cytogenetic data to more comprehensively understand the role genome architecture plays in genome plasticity. We propose adoption of the term 'chromosomics' as an approach encompassing genome sequencing, cytogenetics and cell biology, and present examples of where chromosomics has already led to novel discoveries, such as the sex-determining gene in eutherian mammals. More importantly, we look to the future and the questions that could be answered as we enter into the chromosomics revolution, such as the role of chromosome rearrangements in speciation and the role more rapidly evolving regions of the genome, like centromeres, play in genome plasticity. However, for chromosomics to reach its full potential, we need to address several challenges, particularly the training of a new generation of cytogeneticists, and the commitment to a closer union among the research areas of genomics, cytogenetics, cell biology and bioinformatics. Overcoming these challenges will lead to ground-breaking discoveries in understanding genome evolution and function.
Keywords: centromere; chromosome rearrangements; cytogenetics; evolution; genome biology; genome plasticity; sex chromosomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
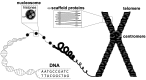

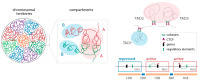
References
-
- Winkler H. Verbreitung und Ursache der Parthenogenesis im Pflanzen-und Tierreiche. Verlag von Gustav Fischer; Jena, Germany: 1920.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

