Ground State Destabilization in Uracil DNA Glycosylase: Let's Not Forget "Tautomeric Strain" in Substrates
- PMID: 31434485
- PMCID: PMC6726543
- DOI: 10.1021/jacs.9b06447
Ground State Destabilization in Uracil DNA Glycosylase: Let's Not Forget "Tautomeric Strain" in Substrates
Abstract
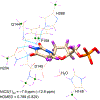
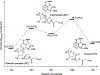
Enzymes like uracil DNA glycosylase (UDG) can achieve ground state destabilization, by polarizing substrates to mimic rare tautomers. On the basis of computed nucleus independent chemical shifts, NICS(1)zz, and harmonic oscillator model of electron delocalization (HOMED) analyses, of quantum mechanics (QM) and quantum mechanics/molecular mechanics (QM/MM) models of the UDG active site, uracil is strongly polarized when bound to UDG and resembles a tautomer >12 kcal/mol higher in energy. Natural resonance theory (NRT) analyses identified a dominant O2 imidate resonance form for residue bound 1-methyl-uracil. This "tautomeric strain" raises the energy of uracil, making uracilate a better than expected leaving group. Computed gas-phase SN2 reactions of free and hydrogen bonded 1-methyl-uracil demonstrate the relationship between the degree of polarization in uracil and the leaving group ability of uracilate.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Biochemical characterization and mutational studies of a thermostable uracil DNA glycosylase from the hyperthermophilic euryarchaeon Thermococcus barophilus Ch5.Int J Biol Macromol. 2019 Aug 1;134:846-855. doi: 10.1016/j.ijbiomac.2019.05.073. Epub 2019 May 14. Int J Biol Macromol. 2019. PMID: 31100400
-
QM/MM Study of the Uracil DNA Glycosylase Reaction Mechanism: A Competition between Asp145 and His148.J Chem Theory Comput. 2019 Aug 13;15(8):4344-4350. doi: 10.1021/acs.jctc.8b01305. Epub 2019 Jul 18. J Chem Theory Comput. 2019. PMID: 31318548
-
Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase.Biochemistry. 1999 Jan 19;38(3):952-63. doi: 10.1021/bi9818669. Biochemistry. 1999. PMID: 9893991
-
Thermodynamic Stability of DNA Duplexes Comprising the Simplest T → dU Substitutions.Biochemistry. 2018 Oct 2;57(39):5666-5671. doi: 10.1021/acs.biochem.8b00676. Epub 2018 Sep 17. Biochemistry. 2018. PMID: 30185020
-
On Prototropy and Bond Length Alternation in Neutral and Ionized Pyrimidine Bases and Their Model Azines in Vacuo.Molecules. 2023 Oct 26;28(21):7282. doi: 10.3390/molecules28217282. Molecules. 2023. PMID: 37959699 Free PMC article. Review.
Cited by
-
Development and application of quantum mechanics/molecular mechanics methods with advanced polarizable potentials.Wiley Interdiscip Rev Comput Mol Sci. 2021 Jul-Aug;11(4):e1515. doi: 10.1002/wcms.1515. Epub 2021 Jan 12. Wiley Interdiscip Rev Comput Mol Sci. 2021. PMID: 34367343 Free PMC article.
References
-
- Jencks WP Catalysis in Chemistry and Enzymology; McGraw-Hill: New York, 1969; p 282.
-
- Menger FM Analysis of ground-state and transition-state effects in enzyme catalysis. Biochemistry 1992, 31, 5368–5373. - PubMed
-
- Savva R; McAuley-Hecht K; Brown T; Pearl L The structural basis of specific base-excision repair by uracil–DNA glycosylase. Nature 1995, 373, 487–493. - PubMed
-
- Mol CD; Arvai AS; Slupphaug G; Kavli B; Alseth I; Krokan HE; Tainer JA Crystal structure of human uracil−DNA glycosylase in complex with a protein inhibitor: Protein mimicry of DNA. Cell 1995, 80, 869–878. - PubMed
-
- Slupphaug G; Mol CD; Kavli B; Arvai AS; Krokan HE; Tainer JA A nucleotide-flipping mechanism from the structure of human uracil–DNA glycosylase bound to DNA. Nature 1996, 384, 87–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

