Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity
- PMID: 31435009
- PMCID: PMC6939392
- DOI: 10.1038/s41586-019-1485-8
Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity
Erratum in
-
Author Correction: Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity.Nature. 2019 Nov;575(7784):E7. doi: 10.1038/s41586-019-1755-5. Nature. 2019. PMID: 31712625
Abstract
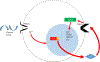
Direct recognition of invading pathogens by innate immune cells is a critical driver of the inflammatory response. However, cells of the innate immune system can also sense their local microenvironment and respond to physiological fluctuations in temperature, pH, oxygen and nutrient availability, which are altered during inflammation. Although cells of the immune system experience force and pressure throughout their life cycle, little is known about how these mechanical processes regulate the immune response. Here we show that cyclical hydrostatic pressure, similar to that experienced by immune cells in the lung, initiates an inflammatory response via the mechanically activated ion channel PIEZO1. Mice lacking PIEZO1 in innate immune cells showed ablated pulmonary inflammation in the context of bacterial infection or fibrotic autoinflammation. Our results reveal an environmental sensory axis that stimulates innate immune cells to mount an inflammatory response, and demonstrate a physiological role for PIEZO1 and mechanosensation in immunity.
Figures


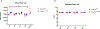
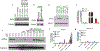





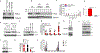

Comment in
-
Pressure regulates immune-cell function.Nature. 2019 Sep;573(7772):41-42. doi: 10.1038/d41586-019-02339-4. Nature. 2019. PMID: 31481766 No abstract available.
-
Monocytes feel the pressure.Nat Rev Immunol. 2019 Oct;19(10):595. doi: 10.1038/s41577-019-0220-x. Nat Rev Immunol. 2019. PMID: 31485035 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

