Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus
- PMID: 31435014
- PMCID: PMC6717508
- DOI: 10.1038/s41586-019-1501-z
Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus
Abstract
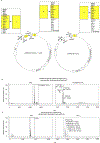
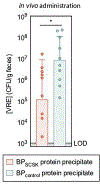
Intestinal commensal bacteria can inhibit dense colonization of the gut by vancomycin-resistant Enterococcus faecium (VRE), a leading cause of hospital-acquired infections1,2. A four-strained consortium of commensal bacteria that contains Blautia producta BPSCSK can reverse antibiotic-induced susceptibility to VRE infection3. Here we show that BPSCSK reduces growth of VRE by secreting a lantibiotic that is similar to the nisin-A produced by Lactococcus lactis. Although the growth of VRE is inhibited by BPSCSK and L. lactis in vitro, only BPSCSK colonizes the colon and reduces VRE density in vivo. In comparison to nisin-A, the BPSCSK lantibiotic has reduced activity against intestinal commensal bacteria. In patients at high risk of VRE infection, high abundance of the lantibiotic gene is associated with reduced density of E. faecium. In germ-free mice transplanted with patient-derived faeces, resistance to VRE colonization correlates with abundance of the lantibiotic gene. Lantibiotic-producing commensal strains of the gastrointestinal tract reduce colonization by VRE and represent potential probiotic agents to re-establish resistance to VRE.
Figures






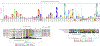





References
-
- Gilmore M, Clewell D, Ike Y & Shankar N Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. (2014). - PubMed
-
- U.S. Centers for Disease Control and Prevention, U. S. D. o. H. a. H. S. Antibiotic resistance threats in the United States, 2013, (U.S. Centers for Disease Control and Prevention, 2013).
Additional References
-
- Montalban-Lopez M, Buivydas A & Kuipers OP in Hydrocarbon and Lipid Microbiology Protocols Springer Protocols Handbooks (eds McGenity T, Timmis K, & Fernandez B. Nogales) (Springer, Berlin, Heidelberg, 2015).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

