Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex
- PMID: 31436533
- PMCID: PMC6721835
- DOI: 10.7554/eLife.48630
Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex
Abstract
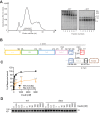
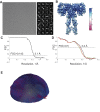
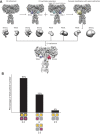
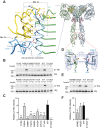
Insulin signaling controls metabolic homeostasis. Here, we report the cryo-EM structure of full-length insulin receptor (IR) and insulin complex in the active state. This structure unexpectedly reveals that maximally four insulins can bind the 'T'-shaped IR dimer at four distinct sites related by 2-fold symmetry. Insulins 1 and 1' bind to sites 1 and 1', formed by L1 of one IR protomer and α-CT and FnIII-1 of the other. Insulins 2 and 2' bind to sites 2 and 2' on FnIII-1 of each protomer. Mutagenesis and cellular assays show that both sites 1 and 2 are required for optimal insulin binding and IR activation. We further identify a homotypic FnIII-2-FnIII-2 interaction in mediating the dimerization of membrane proximal domains in the active IR dimer. Our results indicate that binding of multiple insulins at two distinct types of sites disrupts the autoinhibited apo-IR dimer and stabilizes the active dimer.
Keywords: cryo-EM; human; insulin receptor; molecular biophysics; site 2; structural biology.
© 2019, Uchikawa et al.
Conflict of interest statement
EU, EC, GS, HY, XB No competing interests declared
Figures









References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- RP120717-P2/Cancer Preventation and Research Institute of Texas/International
- I-1944/Welch Foundation/International
- RP160667-P2/Cancer Prevention and Research Institute of Texas/International
- RR160082/Cancer Prevention and Research Institute of Texas/International
- R01 GM136976/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

