Depletion of the Transcriptional Coactivator Amplified in Breast Cancer 1 (AIB1) Uncovers Functionally Distinct Subpopulations in Triple-Negative Breast Cancer
- PMID: 31437536
- PMCID: PMC6706655
- DOI: 10.1016/j.neo.2019.07.001
Depletion of the Transcriptional Coactivator Amplified in Breast Cancer 1 (AIB1) Uncovers Functionally Distinct Subpopulations in Triple-Negative Breast Cancer
Abstract
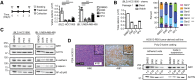
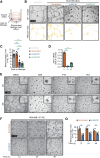
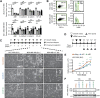
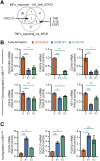
The transcriptional coactivator Amplified in Breast Cancer 1 (AIB1) plays a major role in the progression of hormone and HER2-dependent breast cancers but its role in triple negative breast cancer (TNBC) is undefined. Here, we report that established TNBC cell lines, as well as cells from a TNBC patient-derived xenograft (PDX) that survive chemotherapy treatment in vitro express lower levels of AIB1 protein. The surviving cell population has an impaired tube-formation phenotype when cultured onto basement membrane, a property shared with TNBC cells that survive shRNA-mediated depletion of AIB1 (AIB1LOW cells). DNA analysis by exome sequencing revealed that AIB1LOW cells represent a distinct subpopulation. Consistent with their in vitro phenotype AIB1LOW cells implanted orthotopically generated slower growing tumors with less capacity for pulmonary metastases. Gene expression analysis of cultured cells and tumors revealed that AIB1LOW cells display a distinct expression signature of genes in pro-inflammatory pathways, cell adhesion, proteolysis and tissue remodeling. Interestingly, the presence of this AIB1LOW expression signature in breast cancer specimens is associated with shorter disease free survival of chemotherapy treated patients. We concluded that TNBC cell lines contain heterogeneous populations with differential dependence on AIB1 and that the gene expression pattern of AIB1LOW cells may represent a signature indicative of poor response to chemotherapy in TNBC patients.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







Similar articles
-
Activating Transcription Factor 4 Modulates TGFβ-Induced Aggressiveness in Triple-Negative Breast Cancer via SMAD2/3/4 and mTORC2 Signaling.Clin Cancer Res. 2018 Nov 15;24(22):5697-5709. doi: 10.1158/1078-0432.CCR-17-3125. Epub 2018 Jul 16. Clin Cancer Res. 2018. PMID: 30012564
-
CIB1 depletion impairs cell survival and tumor growth in triple-negative breast cancer.Breast Cancer Res Treat. 2015 Jul;152(2):337-46. doi: 10.1007/s10549-015-3458-4. Epub 2015 Jun 24. Breast Cancer Res Treat. 2015. PMID: 26105795 Free PMC article.
-
Preclinical evaluation of cyclin dependent kinase 11 and casein kinase 2 survival kinases as RNA interference targets for triple negative breast cancer therapy.Breast Cancer Res. 2015;17:19. doi: 10.1186/s13058-015-0524-0. Epub 2015 Feb 11. Breast Cancer Res. 2015. PMID: 25837326 Free PMC article.
-
Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer.Mol Cancer. 2016 Apr 30;15(1):30. doi: 10.1186/s12943-016-0515-5. Mol Cancer. 2016. PMID: 27130446 Free PMC article.
-
The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer.Breast Cancer Res Treat. 2009 Jul;116(2):225-37. doi: 10.1007/s10549-009-0405-2. Epub 2009 May 6. Breast Cancer Res Treat. 2009. PMID: 19418218 Free PMC article. Review.
Cited by
-
SRC-3, a Steroid Receptor Coactivator: Implication in Cancer.Int J Mol Sci. 2021 Apr 30;22(9):4760. doi: 10.3390/ijms22094760. Int J Mol Sci. 2021. PMID: 33946224 Free PMC article. Review.
-
AIB1 is a novel target of the high-risk HPV E6 protein and a biomarker of cervical cancer progression.J Med Virol. 2022 Aug;94(8):3962-3977. doi: 10.1002/jmv.27795. Epub 2022 Apr 27. J Med Virol. 2022. PMID: 35437795 Free PMC article.
References
-
- Wolff AC. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2006;25(1):118–145. - PubMed
-
- Perou CM. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. - PubMed
-
- Nielsen TO Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

