Comparative transcriptomics of early petal development across four diverse species of Aquilegia reveal few genes consistently associated with nectar spur development
- PMID: 31438840
- PMCID: PMC6704642
- DOI: 10.1186/s12864-019-6002-9
Comparative transcriptomics of early petal development across four diverse species of Aquilegia reveal few genes consistently associated with nectar spur development
Abstract
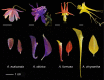
Background: Petal nectar spurs, which facilitate pollination through animal attraction and pollen placement, represent a key innovation promoting diversification in the genus Aquilegia (Ranunculaceae). Identifying the genetic components that contribute to the development of these three-dimensional structures will inform our understanding of the number and types of genetic changes that are involved in the evolution of novel traits. In a prior study, gene expression between two regions of developing petals, the laminar blade and the spur cup, was compared at two developmental stages in the horticultural variety A. coerulea 'Origami'. Several hundred genes were differentially expressed (DE) between the blade and spur at both developmental stages. In order to narrow in on a set of genes crucial to early spur formation, the current study uses RNA sequencing (RNAseq) to conduct comparative expression analyses of petals from five developmental stages between four Aquilegia species, three with morphologically variable nectar spurs, A. sibirica, A. formosa, and A. chrysantha, and one that lacks nectar spurs, A. ecalcarata.
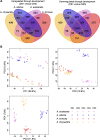
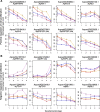
Results: Petal morphology differed increasingly between taxa across the developmental stages assessed, with petals from all four taxa being indistinguishable pre-spur formation at developmental stage 1 (DS1) and highly differentiated by developmental stage 5 (DS5). In all four taxa, genes involved in mitosis were down-regulated over the course of the assessed developmental stages, however, many genes involved in mitotic processes remained expressed at higher levels later in development in the spurred taxa. A total of 690 genes were identified that were consistently DE between the spurred taxa and A. ecalcarata at all five developmental stages. By comparing these genes with those identified as DE between spur and blade tissue in A. coerulea 'Origami', a set of only 35 genes was identified that shows consistent DE between petal samples containing spur tissue versus those without spur tissue.
Conclusions: The results of this study suggest that expression differences in very few loci are associated with the presence and absence of spurs. In general, it appears that the spurless petals of A. ecalcarata cease cell divisions and enter the cell differentiation phase at an earlier developmental time point than those that produce spurs. This much more tractable list of 35 candidates genes will greatly facilitate targeted functional studies to assess the genetic control and evolution of petal spurs in Aquilegia.
Keywords: Aquilegia; Diversification; Evolution; Gene expression; Nectar spur; Petal development; RNAseq.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Filiault DL, Ballerini ES, Mandáková T, Aköz G, Derieg NJ, Schmutz J, Jenkins J, Grimwood J, Shu S, Hayes RD, Hellsten U, Barry K, Yan J, Mihaltcheva S, Karafiátová M, Nizhynska V, Kramer EM, Lysak MA, Hodges SA, Nordborg M. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife. 2018;7:503. doi: 10.7554/eLife.36426. - DOI - PMC - PubMed
-
- Hodges SA. Floral Nectar Spurs and Diversification. Int J Plant Sci. 1997;158(S6):81–8. doi: 10.1086/297508. - DOI
-
- Hodges SA, Fulton M, Yang JY, Whittall JB. Verne Grant and evolutionary studies of Aquilegia. New Phytol. 2003;161(1):113–20. doi: 10.1046/j.1469-8137.2003.00950.x. - DOI

