Hantavirus entry: Perspectives and recent advances
- PMID: 31439149
- PMCID: PMC6881143
- DOI: 10.1016/bs.aivir.2019.07.002
Hantavirus entry: Perspectives and recent advances
Abstract
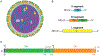
Hantaviruses are important zoonotic pathogens of public health importance that are found on all continents except Antarctica and are associated with hemorrhagic fever with renal syndrome (HFRS) in the Old World and hantavirus pulmonary syndrome (HPS) in the New World. Despite the significant disease burden they cause, no FDA-approved specific therapeutics or vaccines exist against these lethal viruses. The lack of available interventions is largely due to an incomplete understanding of hantavirus pathogenesis and molecular mechanisms of virus replication, including cellular entry. Hantavirus Gn/Gc glycoproteins are the only viral proteins exposed on the surface of virions and are necessary and sufficient to orchestrate virus attachment and entry. In vitro studies have implicated integrins (β1-3), DAF/CD55, and gC1qR as candidate receptors that mediate viral attachment for both Old World and New World hantaviruses. Recently, protocadherin-1 (PCDH1) was demonstrated as a requirement for cellular attachment and entry of New World hantaviruses in vitro and lethal HPS in vivo, making it the first clade-specific host factor to be identified. Attachment of hantavirus particles to cellular receptors induces their internalization by clathrin-mediated, dynamin-independent, or macropinocytosis-like mechanisms, followed by particle trafficking to an endosomal compartment where the fusion of viral and endosomal membranes can occur. Following membrane fusion, which requires cholesterol and acid pH, viral nucleocapsids escape into the cytoplasm and launch genome replication. In this review, we discuss the current mechanistic understanding of hantavirus entry, highlight gaps in our existing knowledge, and suggest areas for future inquiry.
Keywords: Bunyavirus; Hantavirus; Hantavirus pulmonary syndrome; Hemorrhagic fever with renal syndrome; Vaccines; Viral entry; Viral glycoprotein structure; Viral glycoproteins; Viral receptors.
© 2019 Elsevier Inc. All rights reserved.
Figures


References
-
- Aplin JD, Spanswick C, Behzad F, Kimber SJ, Vićovac L, 1996. Integrins beta 5, beta 3 and alpha v are apically distributed in endometrial epithelium. Mol. Hum. Reprod 2, 527–534. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

