A chromatin-based mechanism controls differential regulation of the cytochrome P450 gene Cyp24a1 in renal and non-renal tissues
- PMID: 31439663
- PMCID: PMC6768633
- DOI: 10.1074/jbc.RA119.010173
A chromatin-based mechanism controls differential regulation of the cytochrome P450 gene Cyp24a1 in renal and non-renal tissues
Abstract
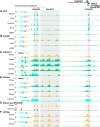
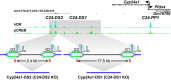
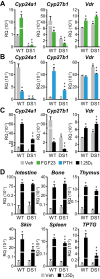
Cytochrome P450 family 27 subfamily B member 1 (CYP27B1) and CYP24A1 function to maintain physiological levels of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) in the kidney. Renal Cyp27b1 and Cyp24a1 expression levels are transcriptionally regulated in a highly reciprocal manner by parathyroid hormone (PTH), fibroblast growth factor 23 (FGF23), and 1,25(OH)2D3 In contrast, Cyp24a1 regulation in nonrenal target cells (NRTCs) is limited to induction by 1,25(OH)2D3 Herein, we used ChIP-Seq analyses of mouse tissues to identify regulatory regions within the Cyp24a1 gene locus. We found an extended region downstream of Cyp24a1 containing a cluster of sites, termed C24-DS1, binding PTH-sensitive cAMP-responsive element-binding protein (CREB) and a cluster termed C24-DS2 binding the vitamin D receptor (VDR). VDR-occupied sites were present in both the kidney and NRTCs, but pCREB sites were occupied only in the kidney. We deleted each segment in the mouse and observed that although the overt phenotypes of both cluster deletions were unremarkable, RNA analysis in the C24-DS1-deleted strain revealed a loss of basal renal Cyp24a1 expression, total resistance to FGF23 and PTH regulation, and secondary suppression of renal Cyp27b1; 1,25(OH)2D3 induction remained unaffected in all tissues. In contrast, loss of the VDR cluster in the C24-DS2-deleted strain did not affect 1,25(OH)2D3 induction of renal Cyp24a1 expression yet reduced but did not eliminate Cyp24a1 responses in NRTCs. We conclude that a chromatin-based mechanism differentially regulates Cyp24a1 in the kidney and NRTCs and is essential for the specific functions of Cyp24a1 in these two tissue types.
Keywords: 1,25(OH)2D3; CRISPR/Cas; ChIP-sequencing (ChIP-Seq); Cyp24a1; Cyp27b1; Cyp27b1-KO; FGF23; cytochrome P450; fibroblast growth factor (FGF); gene regulation; parathyroid hormone (PTH); vitamin D.
© 2019 Meyer et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

