How do cells cope with RNA damage and its consequences?
- PMID: 31439666
- PMCID: PMC6791314
- DOI: 10.1074/jbc.REV119.006513
How do cells cope with RNA damage and its consequences?
Abstract
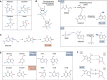
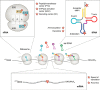
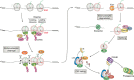
Similar to many other biological molecules, RNA is vulnerable to chemical insults from endogenous and exogenous sources. Noxious agents such as reactive oxygen species or alkylating chemicals have the potential to profoundly affect the chemical properties and hence the function of RNA molecules in the cell. Given the central role of RNA in many fundamental biological processes, including translation and splicing, changes to its chemical composition can have a detrimental impact on cellular fitness, with some evidence suggesting that RNA damage has roles in diseases such as neurodegenerative disorders. We are only just beginning to learn about how cells cope with RNA damage, with recent studies revealing the existence of quality-control processes that are capable of recognizing and degrading or repairing damaged RNA. Here, we begin by reviewing the most abundant types of chemical damage to RNA, including oxidation and alkylation. Focusing on mRNA damage, we then discuss how alterations to this species of RNA affect its function and how cells respond to these challenges to maintain proteostasis. Finally, we briefly discuss how chemical damage to noncoding RNAs such as rRNA, tRNA, small nuclear RNA, and small nucleolar RNA is likely to affect their function.
Keywords: Alzheimer disease; RNA; RNA damage; RNA modification; RNA repair; alkB; alkylation; mRNA surveillance; oxidative stress; quality control; ribosome; stress; translation; ubiquitin.
© 2019 Yan and Zaher.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
References
-
- Mylonas C., and Kouretas D. (1999) Lipid peroxidation and tissue damage. In Vivo 13, 295–309 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

