Variation in Expression of the HECT E3 Ligase UPL3 Modulates LEC2 Levels, Seed Size, and Crop Yields in Brassica napus
- PMID: 31439805
- PMCID: PMC6790077
- DOI: 10.1105/tpc.18.00577
Variation in Expression of the HECT E3 Ligase UPL3 Modulates LEC2 Levels, Seed Size, and Crop Yields in Brassica napus
Abstract
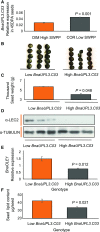
Identifying genetic variation that increases crop yields is a primary objective in plant breeding. We used association analyses of oilseed rape/canola (Brassica napus) accessions to identify genetic variation that influences seed size, lipid content, and final crop yield. Variation in the promoter region of the HECT E3 ligase gene BnaUPL3 C03 made a major contribution to variation in seed weight per pod, with accessions exhibiting high seed weight per pod having lower levels of BnaUPL3 C03 expression. We defined a mechanism in which UPL3 mediated the proteasomal degradation of LEC2, a master transcriptional regulator of seed maturation. Accessions with reduced UPL3 expression had increased LEC2 protein levels, larger seeds, and prolonged expression of lipid biosynthetic genes during seed maturation. Natural variation in BnaUPL3 C03 expression appears not to have been exploited in current B napus breeding lines and could therefore be used as a new approach to maximize future yields in this important oil crop.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures






Comment in
-
Promoting Production: UPL3 Promoter Variation Modulates Seed Size and Crop Yields.Plant Cell. 2019 Oct;31(10):2287-2288. doi: 10.1105/tpc.19.00613. Epub 2019 Aug 22. Plant Cell. 2019. PMID: 31439804 Free PMC article. No abstract available.
References
-
- Andrianov V., Borisjuk N., Pogrebnyak N., Brinker A., Dixon J., Spitsin S., Flynn J., Matyszczuk P., Andryszak K., Laurelli M., Golovkin M., Koprowski H. (2010). Tobacco as a production platform for biofuel: Overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8: 277–287. - PubMed
-
- Badouin H., et al. (2017). The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546: 148–152. - PubMed
-
- Bai X., Huang Y., Hu Y., Liu H., Zhang B., Smaczniak C., Hu G., Han Z., Xing Y. (2017). Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants 3: 885–893. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

