Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland, 2011-2016
- PMID: 31440216
- PMCID: PMC6694793
- DOI: 10.3389/fmicb.2019.01753
Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland, 2011-2016
Erratum in
-
Corrigendum: Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland, 2011-2016.Front Microbiol. 2019 Dec 4;10:2816. doi: 10.3389/fmicb.2019.02816. eCollection 2019. Front Microbiol. 2019. PMID: 31839794 Free PMC article.
Abstract
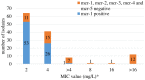
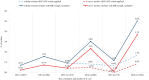
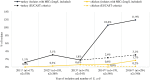
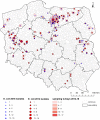
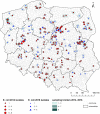
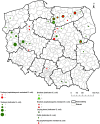
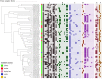
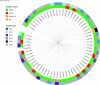
The emergence of plasmid-mediated colistin resistance (mcr genes) threatens the effectiveness of polymyxins, which are last-resort drugs to treat infections by multidrug- and carbapenem-resistant Gram-negative bacteria. Based on the occurrence of colistin resistance the aims of the study were to determine possible resistance mechanisms and then characterize the mcr-positive Escherichia coli. The research used material from the Polish national and EU harmonized antimicrobial resistance (AMR) monitoring programs. A total of 5,878 commensal E. coli from fecal samples of turkeys, chickens, pigs, and cattle collected in 2011-2016 were screened by minimum inhibitory concentration (MIC) determination for the presence of resistance to colistin (R) defined as R > 2 mg/L. Strains with MIC = 2 mg/L isolated in 2014-2016 were also included. A total of 128 isolates were obtained, and most (66.3%) had colistin MIC of 2 mg/L. PCR revealed mcr-1 in 80 (62.5%) isolates recovered from 61 turkeys, 11 broilers, 2 laying hens, 1 pig, and 1 bovine. No other mcr-type genes (including mcr-2 to -5) were detected. Whole-genome sequencing (WGS) of the mcr-1-positive isolates showed high diversity in the multi-locus sequence types (MLST) of E. coli, plasmid replicons, and AMR and virulence genes. Generally mcr-1.1 was detected on the same contig as the IncX4 (76.3%) and IncHI2 (6.3%) replicons. One isolate harbored mcr-1.1 on the chromosome. Various extended-spectrum beta-lactamase (bla SHV-12, bla CTX-M-1, bla CTX-M-15, bla TEM-30, bla TEM-52, and bla TEM-135) and quinolone resistance genes (qnrS1, qnrB19, and chromosomal gyrA, parC, and parE mutations) were present in the mcr-1.1-positive E. coli. A total of 49 sequence types (ST) were identified, ST354, ST359, ST48, and ST617 predominating. One isolate, identified as ST189, belonged to atypical enteropathogenic E. coli. Our findings show that mcr-1.1 has spread widely among production animals in Poland, particularly in turkeys and appears to be transferable mainly by IncX4 and IncHI2 plasmids spread across diverse E. coli lineages. Interestingly, most of these mcr-1-positive E. coli would remain undetected using phenotypic methods with the current epidemiological cut-off value (ECOFF). The appearance and spread of mcr-1 among various animals, but notably in turkeys, might be considered a food chain, and public health hazard.
Keywords: IncHI2; IncX4; WGS; aEPEC; colistin resistance; food animal; mcr-1.
Figures
Similar articles
-
mcr-Colistin Resistance Genes Mobilized by IncX4, IncHI2, and IncI2 Plasmids in Escherichia coli of Pigs and White Stork in Spain.Front Microbiol. 2020 Jan 17;10:3072. doi: 10.3389/fmicb.2019.03072. eCollection 2019. Front Microbiol. 2020. PMID: 32010114 Free PMC article.
-
Emergence and Comparative Genomics Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia coli Carrying mcr-1 in Fennec Fox Imported from Sudan to China.mSphere. 2019 Nov 20;4(6):e00732-19. doi: 10.1128/mSphere.00732-19. mSphere. 2019. PMID: 31748247 Free PMC article.
-
Molecular Epidemiology of mcr-Encoded Colistin Resistance in Enterobacteriaceae From Food-Producing Animals in Italy Revealed Through the EU Harmonized Antimicrobial Resistance Monitoring.Front Microbiol. 2018 Jun 12;9:1217. doi: 10.3389/fmicb.2018.01217. eCollection 2018. Front Microbiol. 2018. PMID: 29951045 Free PMC article.
-
Molecular mechanisms of colistin resistance in Africa: A systematic review of literature.Germs. 2020 Dec 28;10(4):367-379. doi: 10.18683/germs.2020.1229. eCollection 2020 Dec. Germs. 2020. PMID: 33489952 Free PMC article. Review.
-
Emergence and Dissemination of mcr-Carrying Clinically Relevant Salmonella Typhimurium Monophasic Clone ST34.Microorganisms. 2019 Aug 28;7(9):298. doi: 10.3390/microorganisms7090298. Microorganisms. 2019. PMID: 31466338 Free PMC article. Review.
Cited by
-
Dissemination and Comparison of Genetic Determinants of mcr-Mediated Colistin Resistance in Enterobacteriaceae via Retailed Raw Meat Products.Front Microbiol. 2019 Dec 12;10:2824. doi: 10.3389/fmicb.2019.02824. eCollection 2019. Front Microbiol. 2019. PMID: 31921017 Free PMC article.
-
Gain and loss of antibiotic resistant genes in multidrug resistant bacteria: One Health perspective.J Microbiol. 2021 Jun;59(6):535-545. doi: 10.1007/s12275-021-1085-9. Epub 2021 Apr 20. J Microbiol. 2021. PMID: 33877574 Review.
-
Resistance Patterns, mcr-4 and OXA-48 Genes, and Virulence Factors of Escherichia coli from Apennine Chamois Living in Sympatry with Domestic Species, Italy.Animals (Basel). 2022 Jan 6;12(2):129. doi: 10.3390/ani12020129. Animals (Basel). 2022. PMID: 35049753 Free PMC article.
-
Potential sources and characteristic occurrence of mobile colistin resistance (mcr) gene-harbouring bacteria recovered from the poultry sector: a literature synthesis specific to high-income countries.PeerJ. 2021 Oct 5;9:e11606. doi: 10.7717/peerj.11606. eCollection 2021. PeerJ. 2021. PMID: 34707919 Free PMC article.
-
Effect of aromatic oils on the expression of some virulence-associated and antimicrobial resistance genes of Escherichia coli isolated from broilers.J Adv Vet Anim Res. 2022 Jun 26;9(2):191-202. doi: 10.5455/javar.2022.i584. eCollection 2022 Jun. J Adv Vet Anim Res. 2022. PMID: 35891660 Free PMC article.
References
LinkOut - more resources
Full Text Sources

