Phosphorylation Status of Tyrosine 78 Residue Regulates the Nuclear Export and Ubiquitination of Influenza A Virus Nucleoprotein
- PMID: 31440228
- PMCID: PMC6692485
- DOI: 10.3389/fmicb.2019.01816
Phosphorylation Status of Tyrosine 78 Residue Regulates the Nuclear Export and Ubiquitination of Influenza A Virus Nucleoprotein
Abstract
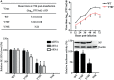
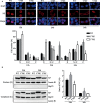
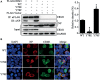
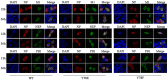
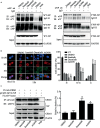
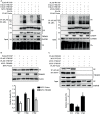
Phosphorylation and dephosphorylation of nucleoprotein (NP) play significant roles in the life cycle of influenza A virus (IAV), and the biological functions of each phosphorylation site on NP are not exactly the same in controlling viral replication. Here, we identified tyrosine 78 residue (Y78) of NP as a novel phosphorylation site by mass spectrometry. Y78 is highly conserved, and the constant NP phosphorylation mimicked by Y78E delayed NP nuclear export through reducing the binding of NP to the cellular export receptor CRM1, and impaired virus growth. Furthermore, the tyrosine kinase inhibitors Dasatinib and AG490 reduced Y78 phosphorylation and accelerated NP nuclear export, suggesting that the Janus and Src kinases-catalyzed Y78 phosphorylation regulated NP nuclear export during viral replication. More importantly, we found that the NP phosphorylation could suppress NP ubiquitination via weakening the interaction between NP and E3 ubiquitin ligase TRIM22, which demonstrated a cross-talk between the phosphorylation and ubiquitination of NP. This study suggests that the phosphorylation status of Y78 regulates IAV replication by inhibiting the nuclear export and ubiquitination of NP. Overall, these findings shed new light on the biological roles of NP phosphorylation, especially its negative role in NP ubiquitination.
Keywords: CRM1; influenza A virus; nuclear export; nucleoprotein; phosphorylation; ubiquitination.
Figures


Similar articles
-
TRIM41-Mediated Ubiquitination of Nucleoprotein Limits Influenza A Virus Infection.J Virol. 2018 Jul 31;92(16):e00905-18. doi: 10.1128/JVI.00905-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29899090 Free PMC article.
-
CNOT4-Mediated Ubiquitination of Influenza A Virus Nucleoprotein Promotes Viral RNA Replication.mBio. 2017 May 23;8(3):e00597-17. doi: 10.1128/mBio.00597-17. mBio. 2017. PMID: 28536288 Free PMC article.
-
Phosphorylation and dephosphorylation of threonine 188 in nucleoprotein is crucial for the replication of influenza A virus.Virology. 2018 Jul;520:30-38. doi: 10.1016/j.virol.2018.05.002. Epub 2018 May 16. Virology. 2018. PMID: 29775781
-
Phosphorylation controls the nuclear-cytoplasmic shuttling of influenza A virus nucleoprotein.J Virol. 2015 Jun;89(11):5822-34. doi: 10.1128/JVI.00015-15. Epub 2015 Mar 18. J Virol. 2015. PMID: 25787277 Free PMC article.
-
Pirh2 restricts influenza A virus replication by modulating short-chain ubiquitination of its nucleoprotein.FASEB J. 2022 Oct;36(10):e22537. doi: 10.1096/fj.202200473R. FASEB J. 2022. PMID: 36070077
Cited by
-
Phosphopeptide enrichment using Phos-tag technology reveals functional phosphorylation of the nucleocapsid protein of SARS-CoV-2.J Proteomics. 2022 Mar 20;255:104501. doi: 10.1016/j.jprot.2022.104501. Epub 2022 Jan 29. J Proteomics. 2022. PMID: 35093569 Free PMC article.
-
Influenza A Virus Uses PSMA2 for Downregulation of the NRF2-Mediated Oxidative Stress Response.J Virol. 2022 Mar 9;96(5):e0199021. doi: 10.1128/jvi.01990-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019712 Free PMC article.
-
Unveiling the role of host kinases at different steps of influenza A virus life cycle.J Virol. 2024 Jan 23;98(1):e0119223. doi: 10.1128/jvi.01192-23. Epub 2024 Jan 4. J Virol. 2024. PMID: 38174932 Free PMC article. Review.
-
Phosphorylation of PB2 at serine 181 restricts viral replication and virulence of the highly pathogenic H5N1 avian influenza virus in mice.Virol Sin. 2024 Feb;39(1):97-112. doi: 10.1016/j.virs.2023.12.003. Epub 2023 Dec 14. Virol Sin. 2024. PMID: 38103645 Free PMC article.
-
PLK3 facilitates replication of swine influenza virus by phosphorylating viral NP protein.Emerg Microbes Infect. 2023 Dec;12(2):2275606. doi: 10.1080/22221751.2023.2275606. Epub 2023 Nov 15. Emerg Microbes Infect. 2023. PMID: 37874309 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

