A Novel Next-Generation Sequencing Approach to Detecting Microsatellite Instability and Pan-Tumor Characterization of 1000 Microsatellite Instability-High Cases in 67,000 Patient Samples
- PMID: 31445211
- PMCID: PMC7807551
- DOI: 10.1016/j.jmoldx.2019.06.011
A Novel Next-Generation Sequencing Approach to Detecting Microsatellite Instability and Pan-Tumor Characterization of 1000 Microsatellite Instability-High Cases in 67,000 Patient Samples
Abstract
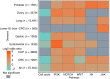
Microsatellite instability (MSI) is an important biomarker for predicting response to immune checkpoint inhibitor therapy, as emphasized by the recent checkpoint inhibitor approval for MSI-high (MSI-H) solid tumors. Herein, we describe and validate a novel method for determining MSI status from a next-generation sequencing comprehensive genomic profiling assay using formalin-fixed, paraffin-embedded samples. This method is 97% (65/67) concordant with current standards, PCR and immunohistochemistry. We further apply this method to >67,000 patient tumor samples to identify genes and pathways that are enriched in MSI-stable or MSI-H tumor groups. Data show that although rare in tumors other than colorectal and endometrial carcinomas, MSI-H samples are present in many tumor types. Furthermore, the large sample set revealed that MSI-H tumors selectively share alterations in genes across multiple common pathways, including WNT, phosphatidylinositol 3-kinase, and NOTCH. Last, MSI is sufficient, but not necessary, for a tumor to have elevated tumor mutation burden. Therefore, MSI can be determined from comprehensive genomic profiling with high accuracy, allowing for efficient MSI-H detection across all tumor types, especially those in which routine use of immunohistochemistry or PCR-based assays would be impractical because of a rare incidence of MSI. MSI-H tumors are enriched in alterations in specific signaling pathways, providing a rationale for investigating directed immune checkpoint inhibitor therapies in combination with pathway-targeted therapies.
Copyright © 2019 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. - PubMed
-
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. - PubMed
-
- Lynch H.T., Snyder C.L., Shaw T.G., Heinen C.D., Hitchins M.P. Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–194. - PubMed
-
- Sinicrope F.A., Foster N.R., Thibodeau S.N., Marsoni S., Monges G., Labianca R., Kim G.P., Yothers G., Allegra C., Moore M.J., Gallinger S., Sargent D.J. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

