Expression Levels of 4 Genes in Colon Tissue Might Be Used to Predict Which Patients Will Enter Endoscopic Remission After Vedolizumab Therapy for Inflammatory Bowel Diseases
- PMID: 31446181
- PMCID: PMC7196933
- DOI: 10.1016/j.cgh.2019.08.030
Expression Levels of 4 Genes in Colon Tissue Might Be Used to Predict Which Patients Will Enter Endoscopic Remission After Vedolizumab Therapy for Inflammatory Bowel Diseases
Abstract
Background & aims: We aimed to identify biomarkers that might be used to predict responses of patients with inflammatory bowel diseases (IBD) to vedolizumab therapy.
Methods: We obtained biopsies from inflamed colon of patients with IBD who began treatment with vedolizumab (n = 31) or tumor necrosis factor (TNF) antagonists (n = 20) and performed RNA-sequencing analyses. We compared gene expression patterns between patients who did and did not enter endoscopic remission (absence of ulcerations at month 6 for patients with Crohn's disease or Mayo endoscopic subscore ≤1 at week 14 for patients with ulcerative colitis) and performed pathway analysis and cell deconvolution for training (n = 20) and validation (n = 11) datasets. Colon biopsies were also analyzed by immunohistochemistry. We validated a baseline gene expression pattern associated with endoscopic remission after vedolizumab therapy using 3 independent datasets (n = 66).
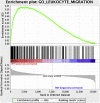
Results: We identified significant differences in expression levels of 44 genes between patients who entered remission after vedolizumab and those who did not; we found significant increases in leukocyte migration in colon tissues from patients who did not enter remission (P < .006). Deconvolution methods identified a significant enrichment of monocytes (P = .005), M1-macrophages (P = .05), and CD4+ T cells (P = .008) in colon tissues from patients who did not enter remission, whereas colon tissues from patients in remission had higher numbers of naïve B cells before treatment (P = .05). Baseline expression levels of PIWIL1, MAATS1, RGS13, and DCHS2 identified patients who did vs did not enter remission with 80% accuracy in the training set and 100% accuracy in validation dataset 1. We validated these findings in the 3 independent datasets by microarray, RNA sequencing and quantitative PCR analysis (P = .003). Expression levels of these 4 genes did not associate with response to anti-TNF agents. We confirmed the presence of proteins encoded by mRNAs using immunohistochemistry.
Conclusions: We identified 4 genes whose baseline expression levels in colon tissues of patients with IBD associate with endoscopic remission after vedolizumab, but not anti-TNF, treatment. We validated this signature in 4 independent datasets and also at the protein level. Studies of these genes might provide insights into the mechanisms of action of vedolizumab.
Keywords: Endoscopic Remisison; IBD; Personalised Medicine; Precision Medicine; Vedolizumab.
Copyright © 2020 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Verstockt B., Verstockt S., Blevi H. TREM-1, the ideal predictive biomarker for endoscopic healing in anti-TNF-treated Crohn's disease patients? Gut. 2019;68:1531–1533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

