Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex
- PMID: 31448322
- PMCID: PMC6691555
- DOI: 10.1126/sciadv.aaw3307
Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex
Abstract
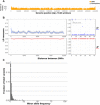
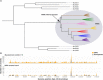
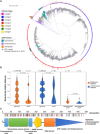
Models on how bacterial lineages differentiate increase our understanding of early bacterial speciation events and the genetic loci involved. Here, we analyze the population genomics events leading to the emergence of the tuberculosis pathogen. The emergence is characterized by a combination of recombination events involving core pathogenesis functions and purifying selection on early diverging loci. We identify the phoR gene, the sensor kinase of a two-component system involved in virulence, as a key functional player subject to pervasive positive selection after the divergence of the Mycobacterium tuberculosis complex from its ancestor. Previous evidence showed that phoR mutations played a central role in the adaptation of the pathogen to different host species. Now, we show that phoR mutations have been under selection during the early spread of human tuberculosis, during later expansions, and in ongoing transmission events. Our results show that linking pathogen evolution across evolutionary and epidemiological time scales points to past and present virulence determinants.
Figures



References
-
- B. J. Shapiro, What microbial population genomics has taught us about speciation, in Population Genomics, M. Polz, O. P. Rajora, Eds. (Springer, 2018).
-
- World Health Organization, Global Tuberculosis Report 2017 (World Health Organization, 2017); www.who.int/tb/publications/global_report/en/.
-
- Comas I., Coscolla M., Luo T., Borrell S., Holt K. E., Kato-Maeda M., Parkhill J., Malla B., Berg S., Thwaites G., Yeboah-Manu D., Bothamley G., Mei J., Wei L., Bentley S., Harris S. R., Niemann S., Diel R., Aseffa A., Gao Q., Young D., Gagneux S., Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182 (2013). - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

