Robustness of Clonogenic Assays as a Biomarker for Cancer Cell Radiosensitivity
- PMID: 31450688
- PMCID: PMC6747107
- DOI: 10.3390/ijms20174148
Robustness of Clonogenic Assays as a Biomarker for Cancer Cell Radiosensitivity
Abstract
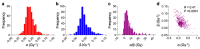
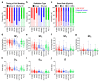
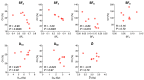
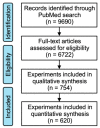
Photon radiation therapy is a major curative treatment for cancer. However, the lack of robust predictive biomarkers for radiosensitivity precludes personalized radiation therapy. Clonogenic assays are the gold standard method for measuring the radiosensitivity of cancer cells. Although a large number of publications describe the use of clonogenic assays to measure cancer cell radiosensitivity, the robustness of results from different studies is unclear. To address this, we conducted a comprehensive detailed literature search of 256 common cancer cell lines and identified the eight cell lines most-frequently examined for photon sensitivity using clonogenic assays. Survival endpoints and experimental parameters from all 620 relevant experiments were compiled and analyzed. We found that the coefficients of variation for SF2 (surviving fraction after 2 Gy irradiation) and for D10 (dose that yields a surviving fraction of 10%) were below 30% for all cell lines, indicating that SF2 and D10 have acceptable inter-assay precision. These data support further analysis of published data on clonogenic assays using SF2 and D10 as survival endpoints, which facilitates robust identification of biological profiles representative of cancer cell sensitivity to photons.
Keywords: cancer; clonogenic assays; precision medicine; radiation therapy; radiosensitivity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. - DOI - PMC - PubMed
-
- Catalogue of Somatic Mutations in Cancer. [(accessed on 24 June 2019)]; Available online: http://cancer.sanger.ac.uk/cosmic.
-
- Steel G.G. End-points for measuring radiation effects on tumours. In: Steel G.G., editor. Basic Clinical Radiobiology. 3rd ed. CRC Press; Boca Raton, FL, USA: 2002. pp. 184–186.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

