Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform
- PMID: 31451149
- PMCID: PMC6777939
- DOI: 10.7554/eLife.46188
Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform
Abstract
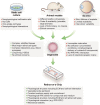
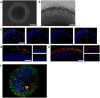
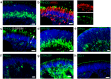
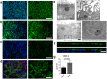
The devastating effects and incurable nature of hereditary and sporadic retinal diseases such as Stargardt disease, age-related macular degeneration or retinitis pigmentosa urgently require the development of new therapeutic strategies. Additionally, a high prevalence of retinal toxicities is becoming more and more an issue of novel targeted therapeutic agents. Ophthalmologic drug development, to date, largely relies on animal models, which often do not provide results that are translatable to human patients. Hence, the establishment of sophisticated human tissue-based in vitro models is of upmost importance. The discovery of self-forming retinal organoids (ROs) derived from human embryonic stem cells (hESCs) or human induced pluripotent stem cells (hiPSCs) is a promising approach to model the complex stratified retinal tissue. Yet, ROs lack vascularization and cannot recapitulate the important physiological interactions of matured photoreceptors and the retinal pigment epithelium (RPE). In this study, we present the retina-on-a-chip (RoC), a novel microphysiological model of the human retina integrating more than seven different essential retinal cell types derived from hiPSCs. It provides vasculature-like perfusion and enables, for the first time, the recapitulation of the interaction of mature photoreceptor segments with RPE in vitro. We show that this interaction enhances the formation of outer segment-like structures and the establishment of in vivo-like physiological processes such as outer segment phagocytosis and calcium dynamics. In addition, we demonstrate the applicability of the RoC for drug testing, by reproducing the retinopathic side-effects of the anti-malaria drug chloroquine and the antibiotic gentamicin. The developed hiPSC-based RoC has the potential to promote drug development and provide new insights into the underlying pathology of retinal diseases.
Keywords: human biology; human induced pulipotent stem cells; medicine; microphysiological systems; none; organ-on-a-chip; regenerative medicine; retinal organoids; retinal pigment epithelial cell; stem cells.
© 2019, Achberger et al.
Conflict of interest statement
KA, CP, JH, SB, JR, JC, MN, VC, LA, WH, NS, KS, MU, SL, PL No competing interests declared
Figures










Comment in
-
Building a better model of the retina.Elife. 2019 Oct 4;8:e51183. doi: 10.7554/eLife.51183. Elife. 2019. PMID: 31584429 Free PMC article.
References
-
- Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Shrike Zhang Y, Shin SR, Calzone G, Annabi N, Shupe TD, Bishop CE, Atala A, Dokmeci MR, Khademhosseini A. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8:014101. doi: 10.1088/1758-5090/8/1/014101. - DOI - PubMed
-
- Blanks JC, Johnson LV. Specific binding of peanut lectin to a class of retinal photoreceptor cells: A species comparison. Investigative Ophthalmology & Visual Science. 1984;25:546–557. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

