Whole-Genome Sequencing To Identify Drivers of Carbapenem-Resistant Klebsiella pneumoniae Transmission within and between Regional Long-Term Acute-Care Hospitals
- PMID: 31451495
- PMCID: PMC6811406
- DOI: 10.1128/AAC.01622-19
Whole-Genome Sequencing To Identify Drivers of Carbapenem-Resistant Klebsiella pneumoniae Transmission within and between Regional Long-Term Acute-Care Hospitals
Abstract
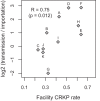
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is an antibiotic resistance threat of the highest priority. Given the limited treatment options for this multidrug-resistant organism (MDRO), there is an urgent need for targeted strategies to prevent transmission. Here, we applied whole-genome sequencing to a comprehensive collection of clinical isolates to reconstruct regional transmission pathways and analyzed this transmission network in the context of statewide patient transfer data and patient-level clinical data to identify drivers of regional transmission. We found that high regional CRKP burdens were due to a small number of regional introductions, with subsequent regional proliferation occurring via patient transfers among health care facilities. While CRKP was predicted to have been imported into each facility multiple times, there was substantial variation in the ratio of intrafacility transmission events per importation, indicating that amplification occurs unevenly across regional facilities. While myriad factors likely influence intrafacility transmission rates, an understudied one is the potential for clinical characteristics of colonized and infected patients to influence their propensity for transmission. Supporting the contribution of high-risk patients to elevated transmission rates, we observed that patients colonized and infected with CRKP in high-transmission facilities had higher rates of carbapenem use, malnutrition, and dialysis and were older. This report highlights the potential for regional infection prevention efforts that are grounded in genomic epidemiology to identify the patients and facilities that make the greatest contribution to regional MDRO prevalence, thereby facilitating the design of precision interventions of maximal impact.
Keywords: carbapenem resistance; epidemiology; genomic epidemiology; long-term acute-care hospitals; regional transmission.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi:10.1128/AAC.45.4.1151-1161.2001. - DOI - PMC - PubMed
-
- Ansari U, Lawsin A, Campbell D, Albrecht V, McAllister G, Bulens S, Walters MS, Jacob JT, Satola SW, Wilson LE, Lynfield R, Vagnone PMS, Janelle SJ, Xavier K, Dumyati G, Hardy D, Phipps EC, Culbreath K, Beldavs Z, Morey K, Kainer MA, Roberts S, Kallen A, Rasheed JK, Karlsson MS. 2017. Molecular characterization of carbapenem-resistant Enterobacteriaceae in the USA, 2011–2015. Open Forum Infect Dis 4:S179. doi:10.1093/ofid/ofx163.328. - DOI
-
- The Wellcome Trust and the UK Department of Health. 2016. Review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. https://amr-review.org/.
-
- Centers for Disease Control and Prevention (CDC). 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA.
-
- World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

