Mutations in the PCNA DNA Polymerase Clamp of Saccharomyces cerevisiae Reveal Complexities of the Cell Cycle and Ploidy on Heterochromatin Assembly
- PMID: 31451562
- PMCID: PMC6781887
- DOI: 10.1534/genetics.119.302452
Mutations in the PCNA DNA Polymerase Clamp of Saccharomyces cerevisiae Reveal Complexities of the Cell Cycle and Ploidy on Heterochromatin Assembly
Abstract
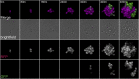
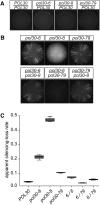
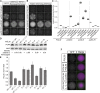
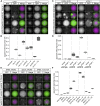
In Saccharomyces cerevisiae, transcriptional silencing at HML and HMR maintains mating-type identity. The repressive chromatin structure at these loci is replicated every cell cycle and must be re-established quickly to prevent transcription of the genes at these loci. Mutations in a component of the replisome, the proliferating cell nuclear antigen (PCNA), encoded by POL30, cause a loss of transcriptional silencing at HMR We used an assay that captures transient losses of silencing at HML and HMR to perform extended genetic analyses of the pol30-6, pol30-8, and pol30-79 alleles. All three alleles destabilized silencing only transiently and only in cycling cells. Whereas pol30-8 caused loss of silencing by disrupting the function of Chromatin Assembly Factor 1, pol30-6 and pol30-79 acted through a separate genetic pathway, but one still dependent on histone chaperones. Surprisingly, the silencing-loss phenotypes of pol30-6 and pol30-79 depended on ploidy, but not on POL30 dosage or mating-type identity. Separately from silencing loss, the pol30-6 and pol30-79 alleles also displayed high levels of mitotic recombination in diploids. These results established that histone trafficking involving PCNA at replication forks is crucial to the maintenance of chromatin state and genome stability during DNA replication. They also raised the possibility that increased ploidy may protect chromatin states when the replisome is perturbed.
Keywords: POL30; intragenic complementation; nucleosome assembly; recombination; transcriptional silencing.
Copyright © 2019 by the Genetics Society of America.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

