Identification of Novel Bacteriophages with Therapeutic Potential That Target Enterococcus faecalis
- PMID: 31451618
- PMCID: PMC6803325
- DOI: 10.1128/IAI.00512-19
Identification of Novel Bacteriophages with Therapeutic Potential That Target Enterococcus faecalis
Abstract
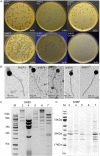
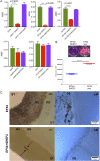
The Gram-positive opportunistic pathogen Enterococcus faecalis is frequently responsible for nosocomial infections in humans and represents one of the most common bacteria isolated from recalcitrant endodontic (root canal) infections. E. faecalis is intrinsically resistant to several antibiotics routinely used in clinical settings (such as cephalosporins and aminoglycosides) and can acquire resistance to vancomycin (vancomycin-resistant enterococci). The resistance of E. faecalis to several classes of antibiotics and its capacity to form biofilms cause serious therapeutic problems. Here, we report the isolation of several bacteriophages that target E. faecalis strains isolated from the oral cavity of patients suffering root canal infections. All phages isolated were Siphoviridae with similar tail lengths (200 to 250 nm) and icosahedral heads. The genome sequences of three isolated phages were highly conserved with the exception of predicted tail protein genes that diverge in sequence, potentially reflecting the host range. The properties of the phage with the broadest host range (SHEF2) were further characterized. We show that this phage requires interaction with components of the major and variant region enterococcal polysaccharide antigen to engage in lytic infection. Finally, we explored the therapeutic potential of this phage and show that it can eradicate E. faecalis biofilms formed in vitro on a standard polystyrene surface but also on a cross-sectional tooth slice model of endodontic infection. We also show that SHEF2 cleared a lethal infection of zebrafish when applied in the circulation. We therefore propose that the phage described here could be used to treat a broad range of antibiotic-resistant E. faecalis infections.
Keywords: bacteriophage; biofilm; capsule; oral microbiology.
Copyright © 2019 Al-Zubidi et al.
Figures




References
-
- Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. doi: 10.1126/science.1080613. - DOI - PubMed
-
- Polidori M, Nuccorini A, Tascini C, Gemignani G, Iapoce R, Leonildi A, Tagliaferri E, Menichetti F. 2011. Vancomycin-resistant Enterococcus faecium (VRE) bacteremia in infective endocarditis successfully treated with combination daptomycin and tigecycline. J Chemother 23:240. doi: 10.1179/joc.2011.23.4.240. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

