Profiling surface proteins on individual exosomes using a proximity barcoding assay
- PMID: 31451692
- PMCID: PMC6710248
- DOI: 10.1038/s41467-019-11486-1
Profiling surface proteins on individual exosomes using a proximity barcoding assay
Abstract
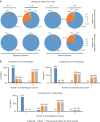
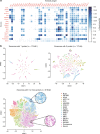
Exosomes have been implicated in numerous biological processes, and they may serve as important disease markers. Surface proteins on exosomes carry information about their tissues of origin. Because of the heterogeneity of exosomes it is desirable to investigate them individually, but this has so far remained impractical. Here, we demonstrate a proximity-dependent barcoding assay to profile surface proteins of individual exosomes using antibody-DNA conjugates and next-generation sequencing. We first validate the method using artificial streptavidin-oligonucleotide complexes, followed by analysis of the variable composition of surface proteins on individual exosomes, derived from human body fluids or cell culture media. Exosomes from different sources are characterized by the presence of specific combinations of surface proteins and their abundance, allowing exosomes to be separately quantified in mixed samples to serve as markers for tissue-specific engagement in disease.
Conflict of interest statement
D.W. has filed a patent application (PCT/SE2014/051133) describing the PBA technique. D.W., U.L., J.Y. and M.K-M hold shares in Vesicode AB commercializing the PBA technology. J.O. is an employee of Vesicode AB. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

