Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin
- PMID: 31452511
- PMCID: PMC6711711
- DOI: 10.7554/eLife.49324
Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin
Abstract
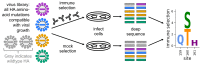
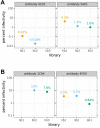
A longstanding question is how influenza virus evolves to escape human immunity, which is polyclonal and can target many distinct epitopes. Here, we map how all amino-acid mutations to influenza's major surface protein affect viral neutralization by polyclonal human sera. The serum of some individuals is so focused that it selects single mutations that reduce viral neutralization by over an order of magnitude. However, different viral mutations escape the sera of different individuals. This individual-to-individual variation in viral escape mutations is not present among ferrets that have been infected just once with a defined viral strain. Our results show how different single mutations help influenza virus escape the immunity of different members of the human population, a phenomenon that could shape viral evolution and disease susceptibility.
Keywords: antigenic drift; deep mutational scanning; evolutionary biology; hemagglutinin; human; infectious disease; influenza virus; microbiology; mutational antigenic profiling; virus.
© 2019, Lee et al.
Conflict of interest statement
JL, RE, SZ, SC, PW, TB, TS, MB, AH, SL, SH, JB No competing interests declared
Figures














Similar articles
-
Sera from Individuals with Narrowly Focused Influenza Virus Antibodies Rapidly Select Viral Escape Mutations In Ovo.J Virol. 2018 Sep 12;92(19):e00859-18. doi: 10.1128/JVI.00859-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30045982 Free PMC article.
-
Complete mapping of viral escape from neutralizing antibodies.PLoS Pathog. 2017 Mar 13;13(3):e1006271. doi: 10.1371/journal.ppat.1006271. eCollection 2017 Mar. PLoS Pathog. 2017. PMID: 28288189 Free PMC article.
-
Deep mutational scanning of H5 hemagglutinin to inform influenza virus surveillance.PLoS Biol. 2024 Nov 12;22(11):e3002916. doi: 10.1371/journal.pbio.3002916. eCollection 2024 Nov. PLoS Biol. 2024. PMID: 39531474 Free PMC article.
-
A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus.J Mol Biol. 2017 Aug 18;429(17):2694-2709. doi: 10.1016/j.jmb.2017.06.015. Epub 2017 Jun 23. J Mol Biol. 2017. PMID: 28648617 Free PMC article. Review.
-
Influenza Hemagglutinin Structures and Antibody Recognition.Cold Spring Harb Perspect Med. 2020 Aug 3;10(8):a038778. doi: 10.1101/cshperspect.a038778. Cold Spring Harb Perspect Med. 2020. PMID: 31871236 Free PMC article. Review.
Cited by
-
Attenuated influenza virions expressing the SARS-CoV-2 receptor-binding domain induce neutralizing antibodies in mice.bioRxiv [Preprint]. 2020 Sep 3:2020.08.12.248823. doi: 10.1101/2020.08.12.248823. bioRxiv. 2020. Update in: Viruses. 2020 Sep 05;12(9):E987. doi: 10.3390/v12090987. PMID: 32817935 Free PMC article. Updated. Preprint.
-
Evolution of Indian Influenza A (H1N1) Hemagglutinin Strains: A Comparative Analysis of the Pandemic Californian HA Strain.Front Mol Biosci. 2023 Mar 16;10:1111869. doi: 10.3389/fmolb.2023.1111869. eCollection 2023. Front Mol Biosci. 2023. PMID: 37006623 Free PMC article.
-
Subdominance in Antibody Responses: Implications for Vaccine Development.Microbiol Mol Biol Rev. 2020 Nov 25;85(1):e00078-20. doi: 10.1128/MMBR.00078-20. Print 2020 Nov 25. Microbiol Mol Biol Rev. 2020. PMID: 33239435 Free PMC article. Review.
-
Antigenic cartography reveals complexities of genetic determinants that lead to antigenic differences among pandemic GII.4 noroviruses.Proc Natl Acad Sci U S A. 2021 Mar 16;118(11):e2015874118. doi: 10.1073/pnas.2015874118. Proc Natl Acad Sci U S A. 2021. PMID: 33836574 Free PMC article.
-
A speed limit on serial strain replacement from original antigenic sin.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2400202121. doi: 10.1073/pnas.2400202121. Epub 2024 Jun 10. Proc Natl Acad Sci U S A. 2024. PMID: 38857397 Free PMC article.
References
-
- Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, Qu X, Lee JH, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, Wilson PC. Immune history profoundly affects broadly protective B cell responses to influenza. Science Translational Medicine. 2015b;7:316ra192. doi: 10.1126/scitranslmed.aad0522. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI108686/NH/NIH HHS/United States
- R01 AI108686/AI/NIAID NIH HHS/United States
- U54GM111274/NH/NIH HHS/United States
- R01 AI127893/AI/NIAID NIH HHS/United States
- HHSN272201400005C/NH/NIH HHS/United States
- F30 AI136326/NH/NIH HHS/United States
- R35 GM119774/GM/NIGMS NIH HHS/United States
- R35 GM119774/NH/NIH HHS/United States
- R01 AI113047/NH/NIH HHS/United States
- R01 AI113047/AI/NIAID NIH HHS/United States
- P01 AI097092/AI/NIAID NIH HHS/United States
- U19 AI117891/NH/NIH HHS/United States
- R01 AI141707/NH/NIH HHS/United States
- S10 OD020069/OD/NIH HHS/United States
- F30 AI136326/AI/NIAID NIH HHS/United States
- T32 GM007266/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

