Development and implementation of a cell-based assay to discover agonists of the nuclear receptor REV-ERBα
- PMID: 31453244
- PMCID: PMC6706147
- DOI: 10.14440/jbm.2018.244
Development and implementation of a cell-based assay to discover agonists of the nuclear receptor REV-ERBα
Abstract
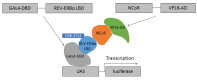
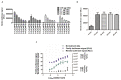
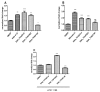
The nuclear receptors are transcription factors involved in the regulation of a variety of physiological processes whose activity can be modulated by binding to relevant small molecule ligands. Their dysfunction has been shown to play a role in disease states such as diabetes, cancer, inflammatory diseases, and hormonal resistance ailments, which makes them interesting targets for drug discovery. The nuclear receptor REV-ERBα is involved in regulating the circadian rhythm and metabolism. Its natural ligand is heme and there is significant interest in identifying novel synthetic modulators to serve as tools to characterize its function and to serve as drugs in treating metabolic disorders. To do so, we established a mammalian cell-based two-hybrid assay system capable of measuring the interaction between REV-ERBα and its co-repressor, nuclear co-repressor 1. This assay was validated to industry standard criteria and was used to screen a subset of the LOPAC®1280 library and 29568 compounds from a diverse compound library. Profiling of the primary hits in a panel of counter and selectivity assays confirmed that REV-ERBα activity can be modulated pharmacologically and chemical scaffolds have been identified for optimization.
Keywords: REV-ERBα; assay development; drug discovery; high throughput screening; luciferase reporter; nuclear receptor.
Conflict of interest statement
Competing interests: The authors have declared that no competing interests exist.
Figures

References
-
- Ottow E, Weinmann H. (2008) Nuclear receptors as drug targets. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA; 498 p.
-
- Burris TP, McCabe ERB. (2001) Nuclear receptors and genetic disease. San Diego: Academic Press; 419 p.
LinkOut - more resources
Full Text Sources
