An Assay to Study Intra-Chromosomal Deletions in Yeast
- PMID: 31454903
- PMCID: PMC6789737
- DOI: 10.3390/mps2030074
An Assay to Study Intra-Chromosomal Deletions in Yeast
Abstract
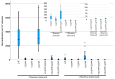
An accurate DNA damage response pathway is critical for the repair of DNA double-strand breaks. Repair may occur by homologous recombination, of which many different sub-pathways have been identified. Some recombination pathways are conservative, meaning that the chromosome sequences are preserved, and others are non-conservative, leading to some alteration of the DNA sequence. We describe an in vivo genetic assay to study non-conservative intra-chromosomal deletions at regions of non-tandem direct repeats in Schizosaccharomyces pombe. This assay can be used to study both spontaneous breaks arising during DNA replication and induced double-strand breaks created with the S. cerevisiae homothallic endonuclease (HO). The preliminary genetic validation of this assay shows that spontaneous breaks require rad52+ but not rad51+, while induced breaks require both genes, in agreement with previous studies. This assay will be useful in the field of DNA damage repair for studying mechanisms of intra-chromosomal deletions.
Keywords: DNA double-strand breaks; Genetic Recombination; Yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

