The mammalian host protein DAP5 facilitates the initial round of translation of Coxsackievirus B3 RNA
- PMID: 31455634
- PMCID: PMC6802510
- DOI: 10.1074/jbc.RA119.009000
The mammalian host protein DAP5 facilitates the initial round of translation of Coxsackievirus B3 RNA
Abstract
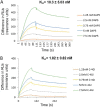
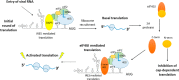
During enteroviral infections, the canonical translation factor eukaryotic translation initiation factor 4 γ I (eIF4GI) is cleaved by viral protease 2A. The resulting C-terminal fragment is recruited by the viral internal ribosome entry site (IRES) for efficient translation of the viral RNA. However, the 2A protease is not present in the viral capsid and is synthesized only after the initial round of translation. This presents the conundrum of how the initial round of translation occurs in the absence of the C-terminal eIF4GI fragment. Interestingly, the host protein DAP5 (also known as p97, eIF4GIII, and eIF4G2), an isoform of eIF4GI, closely resembles the eIF4GI C-terminal fragment produced after 2A protease-mediated cleavage. Using the Coxsackievirus B3 (CVB3) IRES as a model system, here we demonstrate that DAP5, but not the full-length eIF4GI, is required for CVB3 IRES activity for translation of input viral RNA. Additionally, we show that DAP5 is specifically required by type I IRES but not by type II or type III IRES, in which cleavage of eIF4GI has not been observed. We observed that both DAP5 and C-terminal eIF4GI interact with CVB3 IRES in the same region, but DAP5 exhibits a lower affinity for CVB3 IRES compared with the C-terminal eIF4GI fragment. It appears that DAP5 is required for the initial round of viral RNA translation by sustaining a basal level of CVB3 IRES activity. This activity leads to expression of 2A protease and consequent robust CVB3 IRES-mediated translation by the C-terminal eIF4GI fragment.
Keywords: Coxsackievirus B3; DAP5; IRES mediated translation; RNA virus; eukaryotic translation initiation; eukaryotic translation initiation factor 4G (eIF4G); internal initiation of translation; translation; viral protease.
© 2019 Dave et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Etchison D., Milburn S. C., Edery I., Sonenberg N., and Hershey J. W. (1982) Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257, 14806–14810 - PubMed
-
- Gradi A., Svitkin Y. V., Imataka H., and Sonenberg N. (1998) Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U.S.A. 95, 11089–11094 10.1073/pnas.95.19.11089 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

