Interaction of the Ankyrin H Core Effector of Legionella with the Host LARP7 Component of the 7SK snRNP Complex
- PMID: 31455655
- PMCID: PMC6712400
- DOI: 10.1128/mBio.01942-19
Interaction of the Ankyrin H Core Effector of Legionella with the Host LARP7 Component of the 7SK snRNP Complex
Abstract
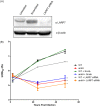
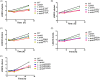
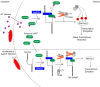
Species of the Legionella genus encode at least 18,000 effector proteins that are translocated through the Dot/Icm type IVB translocation system into macrophages and protist hosts to enable intracellular growth. Eight effectors, including ankyrin H (AnkH), are common to all Legionella species. The AnkH effector is also present in Coxiella and Rickettsiella To date, no pathogenic effectors have ever been described that directly interfere with host cell transcription. We determined that the host nuclear protein La-related protein 7 (LARP7), which is a component of the 7SK small nuclear ribonucleoprotein (snRNP) complex, interacts with AnkH in the host cell nucleus. The AnkH-LARP7 interaction partially impedes interactions of the 7SK snRNP components with LARP7, interfering with transcriptional elongation by polymerase (Pol) II. Consistent with that, our data show AnkH-dependent global reprogramming of transcription of macrophages infected by Legionella pneumophila The crystal structure of AnkH shows that it contains four N-terminal ankyrin repeats, followed by a cysteine protease-like domain and an α-helical C-terminal domain. A substitution within the β-hairpin loop of the third ankyrin repeat results in diminishment of LARP7-AnkH interactions and phenocopies the ankH null mutant defect in intracellular growth. LARP7 knockdown partially suppresses intracellular proliferation of wild-type (WT) bacteria and increases the severity of the defect of the ΔankH mutant, indicating a role for LARP7 in permissiveness of host cells to intracellular bacterial infection. We conclude that the AnkH-LARP7 interaction impedes interaction of LARP7 with 7SK snRNP, which would block transcriptional elongation by Pol II, leading to host global transcriptional reprogramming and permissiveness to L. pneumophilaIMPORTANCE For intracellular pathogens to thrive in host cells, an environment that supports survival and replication needs to be established. L. pneumophila accomplishes this through the activity of the ∼330 effector proteins that are injected into host cells during infection. Effector functions range from hijacking host trafficking pathways to altering host cell machinery, resulting in altered cell biology and innate immunity. One such pathway is the host protein synthesis pathway. Five L. pneumophila effectors have been identified that alter host cell translation, and 2 effectors have been identified that indirectly affect host cell transcription. No pathogenic effectors have been described that directly interfere with host cell transcription. Here we show a direct interaction of the AnkH effector with a host cell transcription complex involved in transcriptional elongation. We identify a novel process by which AnkH interferes with host transcriptional elongation through interference with formation of a functional complex and show that this interference is required for pathogen proliferation.
Keywords: AnkH; Dot/Icm type IVB secretion system; LARP7; Legionella pneumophila; effector functions; transcriptional regulation.
Copyright © 2019 Von Dwingelo et al.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
