CCM1 and CCM2 variants in patients with cerebral cavernous malformation in an ethnically Chinese population in Taiwan
- PMID: 31455779
- PMCID: PMC6712018
- DOI: 10.1038/s41598-019-48448-y
CCM1 and CCM2 variants in patients with cerebral cavernous malformation in an ethnically Chinese population in Taiwan
Abstract
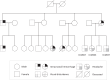
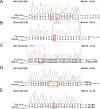
Cerebral cavernous malformation (CCM) is a vascular malformation characterized by clustered enlarged capillary-like channels in the central nervous system. The genes harboring variants in patients with CCM include CCM1/Krev interaction trapped-1, CCM2/MGC4607, and CCM3/programmed cell death protein 10. We aimed to identify pathogenic variants in an ethnic Chinese population in Taiwan. We recruited 95 patients with multiple CCMs or a single lesion with a relevant family history. Sanger sequencing was performed for 41 patients. Variants were identified using sequence alignment tools, and the clinical significance of these variants was determined using American College of Medical Genetics and Genomics standards and guidelines. Several pathogenic variants were found in six patients, including three unrelated patients and three affected members of one family. Two novel pathogenic variants leading to early truncation comprised a deletion variant in exon 18 of CCM1 (c.1846delA; p.Glu617LysfsTer44) and an insertion variant in exon 4 of CCM2 (c.401_402insGCCC; p.Ile136AlafsTer4). One novel pathogenic splice site variant was c.485 + 1G > C at the beginning of intron 8 of CCM1. In this study, we identified novel variants related to CCM in an ethnically Chinese population in Taiwan.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Gunel M, et al. Genetic heterogeneity of inherited cerebral cavernous malformation. Neurosurgery. 1996;38:1265–1271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

