Automatic Brain Tumor Segmentation Based on Cascaded Convolutional Neural Networks With Uncertainty Estimation
- PMID: 31456678
- PMCID: PMC6700294
- DOI: 10.3389/fncom.2019.00056
Automatic Brain Tumor Segmentation Based on Cascaded Convolutional Neural Networks With Uncertainty Estimation
Abstract
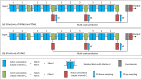
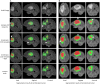
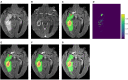
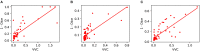
Automatic segmentation of brain tumors from medical images is important for clinical assessment and treatment planning of brain tumors. Recent years have seen an increasing use of convolutional neural networks (CNNs) for this task, but most of them use either 2D networks with relatively low memory requirement while ignoring 3D context, or 3D networks exploiting 3D features while with large memory consumption. In addition, existing methods rarely provide uncertainty information associated with the segmentation result. We propose a cascade of CNNs to segment brain tumors with hierarchical subregions from multi-modal Magnetic Resonance images (MRI), and introduce a 2.5D network that is a trade-off between memory consumption, model complexity and receptive field. In addition, we employ test-time augmentation to achieve improved segmentation accuracy, which also provides voxel-wise and structure-wise uncertainty information of the segmentation result. Experiments with BraTS 2017 dataset showed that our cascaded framework with 2.5D CNNs was one of the top performing methods (second-rank) for the BraTS challenge. We also validated our method with BraTS 2018 dataset and found that test-time augmentation improves brain tumor segmentation accuracy and that the resulting uncertainty information can indicate potential mis-segmentations and help to improve segmentation accuracy.
Keywords: brain tumor segmentation; convolutional neural network; data augmentation; deep learning; uncertainty.
Figures


References
-
- Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., et al. (2016). TensorFlow: A system for large-scale machine learning, in USENIX Symposium on Operating Systems Design and Implementation (Savannah, GA: ), 265–284.
-
- Abdulkadir A., Lienkamp S. S., Brox T., Ronneberger O. (2016). 3D U-Net: Learning dense volumetric segmentation from sparse annotation, in International Conference on Medical Image Computing and Computer Assisted Intervention (Athens: ), 424–432.
-
- Ayhan M. S., Berens P. (2018). Test-time data augmentation for estimation of heteroscedastic aleatoric uncertainty in deep neural networks, in Medical Imaging with Deep Learning (Amsterdam: ), 1–9.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

