The Kinetochore Protein Spc105, a Novel Interaction Partner of LaeA, Regulates Development and Secondary Metabolism in Aspergillus flavus
- PMID: 31456789
- PMCID: PMC6700525
- DOI: 10.3389/fmicb.2019.01881
The Kinetochore Protein Spc105, a Novel Interaction Partner of LaeA, Regulates Development and Secondary Metabolism in Aspergillus flavus
Abstract
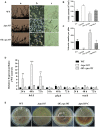
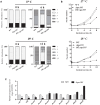
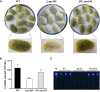
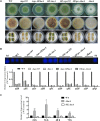
Nuclear protein LaeA is known as the global regulator of secondary metabolism in Aspergillus. LaeA connects with VeA and VelB to form a heterotrimeric complex, which coordinates fungal development and secondary metabolism. Here, we describe a new interaction partner of LaeA, the kinetochore protein Spc105, from the aflatoxin-producing fungus Aspergillus flavus. We showed that in addition to involvement in nuclear division, Spc105 is required for normal conidiophore development and sclerotia production of A. flavus. Moreover, Spc105 positively regulates the production of secondary metabolites such as aflatoxin and kojic acid, and negatively regulates the production of cyclopiazonic acid. Transcriptome analysis of the Δspc105 strain revealed that 23 backbone genes were differentially expressed, corresponding to 19 of the predicted 56 secondary metabolite gene clusters, suggesting a broad regulatory role of Spc105 in secondary metabolism. Notably, the reduced expression of laeA in our transcriptome data led to the discovery of the correlation between Spc105 and LaeA, and double mutant analysis indicated a functional interdependence between Spc105 and LaeA. Further, in vitro and in vivo protein interaction assays revealed that Spc105 interacts directly with the S-adenosylmethionine (SAM)-binding domain of LaeA, and that the leucine zipper motif in Spc105 is required for this interaction. The Spc105-LaeA interaction identified in our study indicates a cooperative interplay of distinct regulators in A. flavus, providing new insights into fungal secondary metabolism regulation networks.
Keywords: Aspergillus flavus; LaeA; Spc105; aflatoxin; protein interaction; secondary metabolism.
Figures






References
-
- Abbas H. K., Accinelli C., Thomas Shier W. (2017). Biological control of aflatoxin contamination in U.S. crops and the use of bioplastic formulations of Aspergillus Flavus biocontrol strains to optimize application Strategies. J. Agric. Food Chem. 65 7081–7087. 10.1021/acs.jafc.7b01452 - DOI - PubMed
LinkOut - more resources
Full Text Sources

