Conserved structural RNA domains in regions coding for cleavage site motifs in hemagglutinin genes of influenza viruses
- PMID: 31456885
- PMCID: PMC6704317
- DOI: 10.1093/ve/vez034
Conserved structural RNA domains in regions coding for cleavage site motifs in hemagglutinin genes of influenza viruses
Abstract
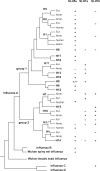
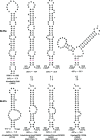
The acquisition of a multibasic cleavage site (MBCS) in the hemagglutinin (HA) glycoprotein is the main determinant of the conversion of low pathogenic avian influenza viruses into highly pathogenic strains, facilitating HA cleavage and virus replication in a broader range of host cells. In nature, substitutions or insertions in HA RNA genomic segments that code for multiple basic amino acids have been observed only in the HA genes of two out of sixteen subtypes circulating in birds, H5 and H7. Given the compatibility of MBCS motifs with HA proteins of numerous subtypes, this selectivity was hypothesized to be determined by the existence of specific motifs in HA RNA, in particular structured domains. In H5 and H7 HA RNAs, predictions of such domains have yielded alternative conserved stem-loop structures with the cleavage site codons in the hairpin loops. Here, potential RNA secondary structures were analyzed in the cleavage site regions of HA segments of influenza viruses of different types and subtypes. H5- and H7-like stem-loop structures were found in all known influenza A virus subtypes and in influenza B and C viruses with homology modeling. Nucleotide covariations supported this conservation to be determined by RNA structural constraints that are stronger in the domain-closing bottom stems as compared to apical parts. The structured character of this region in (sub-)types other than H5 and H7 indicates its functional importance beyond the ability to evolve toward an MBCS responsible for a highly pathogenic phenotype.
Keywords: RNA structure; highly pathogenic avian influenza; influenza virus.
Figures


References
-
- Abolnik C. (2017) ‘Evolution of H5 Highly Pathogenic Avian Influenza: Sequence Data Indicate Stepwise Changes in the Cleavage Site’, Archives of Virology, 162: 2219–30. - PubMed
-
- Alexander D. J. (2007) ‘An Overview of the Epidemiology of Avian Influenza’, Vaccine, 25: 5637–44. - PubMed
-
- Canale A. S. et al. (2018) ‘Synonymous Mutations at the Beginning of the Influenza A Virus Hemagglutinin Gene Impact Experimental Fitness’, Journal of Molecular Biology, 430: 1098–115. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

