Survey of ribose ring pucker of signaling nucleosides and nucleotides
- PMID: 31460850
- PMCID: PMC7047539
- DOI: 10.1080/15257770.2019.1658115
Survey of ribose ring pucker of signaling nucleosides and nucleotides
Abstract
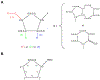
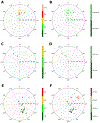
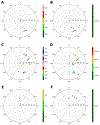
The ribose of protein-bound nucleosides and nucleotides displays preferred conformations (usually either North or South), which can be exploited to design enhanced analogs having chemically fixed conformations. We introduce a computational protocol for assembling data from the protein database (PDB) on the ribose and ribose-like conformation of small molecule ligands when complexed with purinergic signaling proteins (including receptors, enzymes and transporters, and related intracellular pathways). Some targets prefer exclusively North (adenosine and P2Y1 receptors, CD73, adenosine kinase ATP/ADP-binding site, adenosine deaminase), others prefer South (P2Y12 receptor, E-NTPDase2) or East (adenosine kinase substrates), while others (P2XRs) allow various conformations.
Keywords: G protein-coupled receptor; Nucleoside; enzyme; nucleotide; transporter.
Conflict of interest statement
Declaration of Interest Statement:
None.
Figures
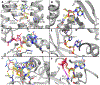
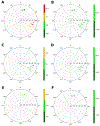
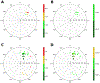
Similar articles
-
Action of nucleosides and nucleotides at 7 transmembrane-spanning receptors.Nucleosides Nucleotides Nucleic Acids. 2006;25(12):1425-36. doi: 10.1080/15257770600919027. Nucleosides Nucleotides Nucleic Acids. 2006. PMID: 17067963 Free PMC article. Review.
-
The nucleoside transport proteins, NupC and NupG, from Escherichia coli: specific structural motifs necessary for the binding of ligands.Org Biomol Chem. 2005 Feb 7;3(3):462-70. doi: 10.1039/b414739a. Epub 2005 Jan 10. Org Biomol Chem. 2005. PMID: 15678184
-
Synthesis of ethyl (1S,2R,3S,4S,5S)-2,3-O-(isopropylidene)-4-hydroxy-bicyclo[3.1.0]hexane-carboxylate from L-ribose: a versatile chiral synthon for preparation of adenosine and P2 receptor ligands.Nucleosides Nucleotides Nucleic Acids. 2008 Mar;27(3):279-91. doi: 10.1080/15257770701845253. Nucleosides Nucleotides Nucleic Acids. 2008. PMID: 18260011 Free PMC article.
-
Ribose modified nucleosides and nucleotides as ligands for purine receptors.Nucleosides Nucleotides Nucleic Acids. 2001 Apr-Jul;20(4-7):333-41. doi: 10.1081/NCN-100002305. Nucleosides Nucleotides Nucleic Acids. 2001. PMID: 11563046 Free PMC article. Review.
-
Stereochemical studies on nucleic acid analogues. I. Conformations of alpha-nucleosides and alpha-nucleotides: interconversion of sugar puckers via O4'-exo.Biopolymers. 1992 Mar;32(3):249-69. doi: 10.1002/bip.360320306. Biopolymers. 1992. PMID: 1581546
Cited by
-
Synthesis and Effect of Conformationally Locked Carbocyclic Guanine Nucleotides on Dynamin.Biomolecules. 2022 Apr 16;12(4):584. doi: 10.3390/biom12040584. Biomolecules. 2022. PMID: 35454173 Free PMC article.
-
Structure-Activity Relationship of 3-Methylcytidine-5'-α,β-methylenediphosphates as CD73 Inhibitors.J Med Chem. 2022 Feb 10;65(3):2409-2433. doi: 10.1021/acs.jmedchem.1c01852. Epub 2022 Jan 26. J Med Chem. 2022. PMID: 35080883 Free PMC article.
-
Expanding the repertoire of methanocarba nucleosides from purinergic signaling to diverse targets.RSC Med Chem. 2021 Jul 13;12(11):1808-1825. doi: 10.1039/d1md00167a. eCollection 2021 Nov 17. RSC Med Chem. 2021. PMID: 34825182 Free PMC article. Review.
-
Purinergic Signaling: Impact of GPCR Structures on Rational Drug Design.ChemMedChem. 2020 Nov 4;15(21):1958-1973. doi: 10.1002/cmdc.202000465. Epub 2020 Sep 18. ChemMedChem. 2020. PMID: 32803849 Free PMC article. Review.
-
2-Substituted (N)-Methanocarba A3 Adenosine Receptor Agonists: In Silico, In Vitro, and In Vivo Characterization.ACS Pharmacol Transl Sci. 2024 Jun 6;7(7):2154-2173. doi: 10.1021/acsptsci.4c00223. eCollection 2024 Jul 12. ACS Pharmacol Transl Sci. 2024. PMID: 39022354 Free PMC article.
References
-
- Linden J; Koch-Nolte F; Dahl G Purine Release, Metabolism, and Signaling in the Inflammatory Response. Annu. Rev. Immunol, 2019, 37, 325–47. - PubMed
-
- Yegutkin GG Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 473–497. - PubMed
-
- Musheshe N; Schmidt M; Zaccolo M cAMP: From long-range second messenger to nanodomain signalling. Trends Pharmacol. Sci, 2018, 39, 209–222. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
