Molecular regulation of human skeletal muscle protein synthesis in response to exercise and nutrients: a compass for overcoming age-related anabolic resistance
- PMID: 31461340
- PMCID: PMC6962519
- DOI: 10.1152/ajpcell.00209.2019
Molecular regulation of human skeletal muscle protein synthesis in response to exercise and nutrients: a compass for overcoming age-related anabolic resistance
Abstract
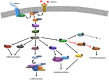
Skeletal muscle mass, a strong predictor of longevity and health in humans, is determined by the balance of two cellular processes, muscle protein synthesis (MPS) and muscle protein breakdown. MPS seems to be particularly sensitive to changes in mechanical load and/or nutritional status; therefore, much research has focused on understanding the molecular mechanisms that underpin this cellular process. Furthermore, older individuals display an attenuated MPS response to anabolic stimuli, termed anabolic resistance, which has a negative impact on muscle mass and function, as well as quality of life. Therefore, an understanding of which, if any, molecular mechanisms contribute to anabolic resistance of MPS is of vital importance in formulation of therapeutic interventions for such populations. This review summarizes the current knowledge of the mechanisms that underpin MPS, which are broadly divided into mechanistic target of rapamycin complex 1 (mTORC1)-dependent, mTORC1-independent, and ribosomal biogenesis-related, and describes the evidence that shows how they are regulated by anabolic stimuli (exercise and/or nutrition) in healthy human skeletal muscle. This review also summarizes evidence regarding which of these mechanisms may be implicated in age-related skeletal muscle anabolic resistance and provides recommendations for future avenues of research that can expand our knowledge of this area.
Keywords: ERK1/2; anabolic resistance; mTORC1; muscle protein synthesis.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures


References
-
- Abou Sawan S, van Vliet S, Parel JT, Beals JW, Mazzulla M, West DWD, Philp A, Li Z, Paluska SA, Burd NA, Moore DR. Translocation and protein complex co-localization of mTOR is associated with postprandial myofibrillar protein synthesis at rest and after endurance exercise. Physiol Rep 6: e13628, 2018. doi: 10.14814/phy2.13628. - DOI - PMC - PubMed
-
- Apró W, Moberg M, Hamilton DL, Ekblom B, Rooyackers O, Holmberg HC, Blomstrand E. Leucine does not affect mechanistic target of rapamycin complex 1 assembly but is required for maximal ribosomal protein S6 kinase 1 activity in human skeletal muscle following resistance exercise. FASEB J 29: 4358–4373, 2015. doi: 10.1096/fj.15-273474. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

