Molecular Portraits of Early Rheumatoid Arthritis Identify Clinical and Treatment Response Phenotypes
- PMID: 31461658
- PMCID: PMC6718830
- DOI: 10.1016/j.celrep.2019.07.091
Molecular Portraits of Early Rheumatoid Arthritis Identify Clinical and Treatment Response Phenotypes
Abstract
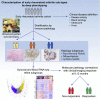
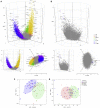
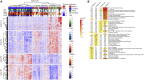
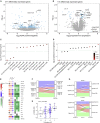
There is a current imperative to unravel the hierarchy of molecular pathways that drive the transition of early to established disease in rheumatoid arthritis (RA). Herein, we report a comprehensive RNA sequencing analysis of the molecular pathways that drive early RA progression in the disease tissue (synovium), comparing matched peripheral blood RNA-seq in a large cohort of early treatment-naive patients, namely, the Pathobiology of Early Arthritis Cohort (PEAC). We developed a data exploration website (https://peac.hpc.qmul.ac.uk/) to dissect gene signatures across synovial and blood compartments, integrated with deep phenotypic profiling. We identified transcriptional subgroups in synovium linked to three distinct pathotypes: fibroblastic pauci-immune pathotype, macrophage-rich diffuse-myeloid pathotype, and a lympho-myeloid pathotype characterized by infiltration of lymphocytes and myeloid cells. This is suggestive of divergent pathogenic pathways or activation disease states. Pro-myeloid inflammatory synovial gene signatures correlated with clinical response to initial drug therapy, whereas plasma cell genes identified a poor prognosis subgroup with progressive structural damage.
Keywords: PEAC; Pathobiology of Early Arthritis Cohort study; RNA sequencing; ectopic lymphoid structures; lymphoid neogenesis; personalized medicine; rheumatoid arthritis; synovial biopsy; transcriptomics.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Badot V., Galant C., Nzeusseu Toukap A., Theate I., Maudoux A.L., Van den Eynde B.J., Durez P., Houssiau F.A., Lauwerys B.R. Gene expression profiling in the synovium identifies a predictive signature of absence of response to adalimumab therapy in rheumatoid arthritis. Arthritis Res. Ther. 2009;11:R57. - PMC - PubMed
-
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. - PubMed
-
- Bresnihan B., Pontifex E., Thurlings R.M., Vinkenoog M., El-Gabalawy H., Fearon U., Fitzgerald O., Gerlag D.M., Rooney T., van de Sande M.G. Synovial tissue sublining CD68 expression is a biomarker of therapeutic response in rheumatoid arthritis clinical trials: consistency across centers. J. Rheumatol. 2009;36:1800–1802. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

