Phylodynamics of Influenza A/H1N1pdm09 in India Reveals Circulation Patterns and Increased Selection for Clade 6b Residues and Other High Mortality Mutants
- PMID: 31462006
- PMCID: PMC6783925
- DOI: 10.3390/v11090791
Phylodynamics of Influenza A/H1N1pdm09 in India Reveals Circulation Patterns and Increased Selection for Clade 6b Residues and Other High Mortality Mutants
Abstract
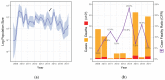
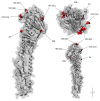
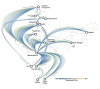
The clinical severity and observed case fatality ratio of influenza A/H1N1pdm09 in India, particularly in 2015 and 2017 far exceeds current global estimates. Reasons for these frequent and severe epidemic waves remain unclear. We used Bayesian phylodynamic methods to uncover possible genetic explanations for this, while also identifying the transmission dynamics of A/H1N1pdm09 between 2009 and 2017 to inform future public health interventions. We reveal a disproportionate selection at haemagglutinin residue positions associated with increased morbidity and mortality in India such as position 222 and clade 6B characteristic residues, relative to equivalent isolates circulating globally. We also identify for the first time, increased selection at position 186 as potentially explaining the severity of recent A/H1N1pdm09 epidemics in India. We reveal national routes of A/H1N1pdm09 transmission, identifying Maharashtra as the most important state for the spread throughout India, while quantifying climactic, ecological, and transport factors as drivers of within-country transmission. Together these results have important implications for future A/H1N1pdm09 surveillance and control within India, but also for epidemic and pandemic risk prediction around the world.
Keywords: India; Influenza; phylogenetics; public health.
Conflict of interest statement
Author C.R.M. has sat on advisory boards for GlaxoSmithKline (GSK), Seqirus, Sanofi and Pfizer and has received funding or in-kind support for investigator-driven research from GSK, Seqirus, Sanofi, Wyeth, and Pfizer unrelated to this study. All other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Genetic characterization and diversity of circulating influenza A/H1N1pdm09 viruses isolated in Jeddah, Saudi Arabia between 2014 and 2015.Arch Virol. 2018 May;163(5):1219-1230. doi: 10.1007/s00705-018-3732-y. Epub 2018 Feb 2. Arch Virol. 2018. PMID: 29396684
-
Evolutionary dynamics of influenza A/H1N1 virus circulating in India from 2011 to 2021.Infect Genet Evol. 2023 Jun;110:105424. doi: 10.1016/j.meegid.2023.105424. Epub 2023 Mar 11. Infect Genet Evol. 2023. PMID: 36913995
-
Genetic diversity of HA1 domain of hemagglutinin gene of pandemic influenza H1N1pdm09 viruses in New Delhi, India.J Med Virol. 2012 Mar;84(3):386-93. doi: 10.1002/jmv.23205. J Med Virol. 2012. PMID: 22246823
-
Molecular characterization of circulating pandemic strains of influenza A virus during 2012 to 2013 in Lucknow (India).J Med Virol. 2014 Dec;86(12):2134-41. doi: 10.1002/jmv.23946. Epub 2014 Apr 29. J Med Virol. 2014. PMID: 24777528
-
Resurrected pandemic influenza viruses.Annu Rev Microbiol. 2009;63:79-98. doi: 10.1146/annurev.micro.091208.073359. Annu Rev Microbiol. 2009. PMID: 19385726 Review.
Cited by
-
Special Issue "Emerging Viruses: Surveillance, Prevention, Evolution, and Control".Viruses. 2020 Mar 11;12(3):306. doi: 10.3390/v12030306. Viruses. 2020. PMID: 32168932 Free PMC article.
-
Molecular Phylogenesis and Spatiotemporal Spread of SARS-CoV-2 in Southeast Asia.Front Public Health. 2021 Jul 30;9:685315. doi: 10.3389/fpubh.2021.685315. eCollection 2021. Front Public Health. 2021. PMID: 34395364 Free PMC article.
-
Modelling the impact of a smallpox attack in India and influence of disease control measures.BMJ Open. 2020 Dec 13;10(12):e038480. doi: 10.1136/bmjopen-2020-038480. BMJ Open. 2020. PMID: 33318109 Free PMC article.
-
In silico analysis and molecular characterization of Influenza A (H1N1) pdm09 virus circulating and causing major outbreaks in central India, 2009-2019.Iran J Microbiol. 2020 Oct;12(5):483-494. doi: 10.18502/ijm.v12i5.4611. Iran J Microbiol. 2020. PMID: 33604005 Free PMC article.
-
Evolution of Indian Influenza A (H1N1) Hemagglutinin Strains: A Comparative Analysis of the Pandemic Californian HA Strain.Front Mol Biosci. 2023 Mar 16;10:1111869. doi: 10.3389/fmolb.2023.1111869. eCollection 2023. Front Mol Biosci. 2023. PMID: 37006623 Free PMC article.
References
-
- Webb S.A.R., Pettilä V., Seppelt I., Bellomo R., Bailey M., Cooper D.J., Cretikos M., Davies A.R., Finfer S., Harrigan P.W.J., et al. Critical Care Services and 2009 H1N1 Influenza in Australia and New Zealand. N. Engl. J. Med. 2009;361:1925–1934. - PubMed
-
- Mitchell R., Ogunremi T., Astrakianakis G., Bryce E., Gervais R., Gravel D., Johnston L., LeDuc S., Roth V., Taylor G., et al. Impact of the 2009 influenza A (H1N1) pandemic on Canadian health care workers: A survey on vaccination, illness, absenteeism, and personal protective equipment. Am. J. Infect. Control. 2012;40:611–616. doi: 10.1016/j.ajic.2012.01.011. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

