Whole-genome sequencing reveals distinct genetic bases for insulinomas and non-functional pancreatic neuroendocrine tumours: leading to a new classification system
- PMID: 31462556
- PMCID: PMC7229893
- DOI: 10.1136/gutjnl-2018-317233
Whole-genome sequencing reveals distinct genetic bases for insulinomas and non-functional pancreatic neuroendocrine tumours: leading to a new classification system
Abstract
Objective: Insulinomas and non-functional pancreatic neuroendocrine tumours (NF-PanNETs) have distinctive clinical presentations but share similar pathological features. Their genetic bases have not been comprehensively compared. Herein, we used whole-genome/whole-exome sequencing (WGS/WES) to identify genetic differences between insulinomas and NF-PanNETs.
Design: The mutational profiles and copy-number variation (CNV) patterns of 211 PanNETs, including 84 insulinomas and 127 NF-PanNETs, were obtained from WGS/WES data provided by Peking Union Medical College Hospital and the International Cancer Genome Consortium. Insulinoma RNA sequencing and immunohistochemistry data were assayed.
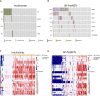
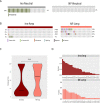
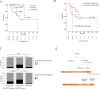
Results: PanNETs were categorised based on CNV patterns: amplification, copy neutral and deletion. Insulinomas had CNV amplifications and copy neutral and lacked CNV deletions. CNV-neutral insulinomas exhibited an elevated rate of YY1 mutations. In contrast, NF-PanNETs had all three CNV patterns, and NF-PanNETs with CNV deletions had a high rate of loss-of-function mutations of tumour suppressor genes. NF-PanNETs with CNV alterations (amplification and deletion) had an elevated risk of relapse, and additional DAXX/ATRX mutations could predict an increased relapse risk in the first 2-year period.
Conclusion: These WGS/WES data allowed a comprehensive assessment of genetic differences between insulinomas and NF-PanNETs, reclassifying these tumours into novel molecular subtypes. We also proposed a novel relapse risk stratification system using CNV patterns and DAXX/ATRX mutations.
Keywords: gene mutation; neuroendocrine tumours; pancreas.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
