LOXL2-mediated H3K4 oxidation reduces chromatin accessibility in triple-negative breast cancer cells
- PMID: 31462706
- PMCID: PMC6937214
- DOI: 10.1038/s41388-019-0969-1
LOXL2-mediated H3K4 oxidation reduces chromatin accessibility in triple-negative breast cancer cells
Abstract
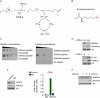
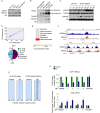
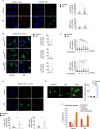
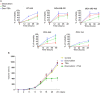
Oxidation of H3 at lysine 4 (H3K4ox) by lysyl oxidase-like 2 (LOXL2) generates an H3 modification with an unknown physiological function. We find that LOXL2 and H3K4ox are higher in triple-negative breast cancer (TNBC) cell lines and patient-derived xenografts (PDXs) than those from other breast cancer subtypes. ChIP-seq revealed that H3K4ox is located primarily in heterochromatin, where it is involved in chromatin compaction. Knocking down LOXL2 reduces H3K4ox levels and causes chromatin decompaction, resulting in a sustained activation of the DNA damage response (DDR) and increased susceptibility to anticancer agents. This critical role that LOXL2 and oxidized H3 play in chromatin compaction and DDR suggests that functionally targeting LOXL2 could be a way to sensitize TNBC cells to conventional therapy.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Williamson PR, Kagan HM. Reaction pathway of bovine aortic lysyl oxidase. J Biol Chem. 1986;261:9477–82. - PubMed
-
- Herranz N, Dave N, Millanes-Romero A, Pascual-Reguant L, Morey L, Diaz VM, et al. Lysyl oxidasE−like 2 (LOXL2) oxidizes trimethylated lysine 4 in histone H3. FEBS J. 2016;283:4263–73. - PubMed
-
- Iturbide A, Garcia de Herreros A, Peiro S. A new role for LOX and LOXL2 proteins in transcription regulation. FEBS J. 2015;282:1768–73. - PubMed
-
- Iturbide A, Pascual-Reguant L, Fargas L, Cebria JP, Alsina B, Garcia de Herreros A, et al. LOXL2 oxidizes methylated TAF10 and controls TFIID-dependent genes during neural progenitor differentiation. Mol Cell. 2015;58:755–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

