Metastatic-niche labelling reveals parenchymal cells with stem features
- PMID: 31462798
- PMCID: PMC6797499
- DOI: 10.1038/s41586-019-1487-6
Metastatic-niche labelling reveals parenchymal cells with stem features
Erratum in
-
Author Correction: Metastatic-niche labelling reveals parenchymal cells with stem features.Nature. 2019 Nov;575(7784):E8. doi: 10.1038/s41586-019-1697-y. Nature. 2019. PMID: 31719688
Abstract
Direct investigation of the early cellular changes induced by metastatic cells within the surrounding tissue remains a challenge. Here we present a system in which metastatic cancer cells release a cell-penetrating fluorescent protein, which is taken up by neighbouring cells and enables spatial identification of the local metastatic cellular environment. Using this system, tissue cells with low representation in the metastatic niche can be identified and characterized within the bulk tissue. To highlight its potential, we applied this strategy to study the cellular environment of metastatic breast cancer cells in the lung. We report the presence of cancer-associated parenchymal cells, which exhibit stem-cell-like features, expression of lung progenitor markers, multi-lineage differentiation potential and self-renewal activity. In ex vivo assays, lung epithelial cells acquire a cancer-associated parenchymal-cell-like phenotype when co-cultured with cancer cells and support their growth. These results highlight the potential of this method as a platform for new discoveries.
Figures

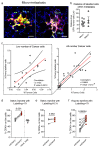












Comment in
-
Cells tagged near an early spread of cancer.Nature. 2019 Aug;572(7771):589-590. doi: 10.1038/d41586-019-02399-6. Nature. 2019. PMID: 31462800 No abstract available.
-
Getting to know the neighbourhood.Nat Rev Cancer. 2019 Nov;19(11):605. doi: 10.1038/s41568-019-0211-z. Nat Rev Cancer. 2019. PMID: 31548603 No abstract available.
-
In vivo labelling system to study cell neighbourhoods.Nat Rev Cancer. 2022 Dec;22(12):661. doi: 10.1038/s41568-022-00521-x. Nat Rev Cancer. 2022. PMID: 36175643 No abstract available.
References
-
- Hanahan D, Coussens LM. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. CCELL. 2012;21:309–322. - PubMed
-
- Barash S, Wang W, Shi Y. Human secretory signal peptide description by hidden Markov model and generation of a strong artificial signal peptide for secreted protein expression. Biochemical and Biophysical Research Communications. 2002;294:835–842. - PubMed
-
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Meth. 2005;2:905–909. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

